2. 河南省农业科学院动物免疫学重点实验室, 农业部动物免疫学重点实验室, 河南省动物免疫学重点实验室, 郑州 450002;
3. 河南省农业科学院, 中英禽病国际研究中心, 郑州 450002;
4. 河南农业大学牧医工程学院, 郑州 450002
2. Key Laboratory of Animal Immunology, Ministry of Agriculture & Henan Provincial Key Laboratory of Animal Immunology, Henan Academy of Agricultural Sciences, Zhengzhou 450002, China;
3. UK-China Centre of Excellence for Research on Avian Diseases, Henan Academy of Agricultural Sciences, Zhengzhou 450002, China;
4. College of Animal Science and Veterinary Medicine, Henan Agricultural University, Zhengzhou 450002, China
马立克病(Marek’s disease,MD)是由鸡马立克病病毒(Marek’s disease virus,MDV)感染鸡引发的一种重要的免疫抑制病及肿瘤病[1]。MD是最早使用疫苗成功预防肿瘤发生的一种肿瘤病,近年来随着疫苗免疫压力增强,MDV的毒力不断增强,在疫苗免疫鸡群中仍时有MD病例发生,特超强毒株可能已突破CVI988/Rispens疫苗的免疫保护,给全球养禽业造成严重经济损失[2]。
CRISPR/Cas9基因编辑技术自2013年首次成功应用于人类免疫缺陷病毒基因编辑以来[3],短短数年间已被广泛应用于DNA病毒的基因编辑,如单纯疱疹病毒、腺病毒、伪狂犬病病毒、牛痘病毒、爱泼斯坦-巴尔病毒、豚鼠巨细胞病毒、鸭肠炎病毒、禽腺病毒4型以及非洲猪瘟病毒等[4-15]。已被编辑的病毒大多属于疱疹病毒。作为α-疱疹病毒的重要成员,MDV是目前已知致瘤性最强的一种,被认为是研究病毒致病及致瘤机制的理想模型[16]。
MDV基因组为线性双股DNA,全长约180 kb[17]。meq是公认的主要致瘤基因,在MD肿瘤发生中具有重要作用[18-19],同时,它也是具有重要进化意义的毒力基因,在强毒株和疫苗毒株之间具有明显的序列差异性。因此,特异识别不同毒力毒株MEQ蛋白的单克隆抗体,可用于MDV感染与免疫的鉴别诊断。本文以CVI988/Rispens的meq基因为靶点,利用CRISPR/Cas9基因编辑技术构建meq基因编辑的缺失毒株,以期为后续差异筛选和鉴定抗MD疫苗株MEQ蛋白的单抗及鉴别诊断研究奠定重要基础。
1 材料与方法 1.1 细胞、毒株、质粒与抗体鸡胚成纤维细胞(CEF)用9~10日龄SPF鸡胚制备;CVI988/Rispens液氮苗购自梅里亚公司;pX459 v2.0质粒和兔抗MDV-MEQ多抗由英国Pirbright研究所Venugopal Nair教授馈赠;MDV-gB单抗由山东农业大学崔治中教授馈赠。
1.2 主要试剂与设备M199培养基、Opti-MEM培养基和胎牛血清(FBS)均购自Gibco公司;0.25%胰酶、双抗、TPB、1 mol·L-1 Tris-HCl(pH 8.0)、0.5 mol·L-1 EDTA(pH 8.0)和蛋白酶K溶液(10 mg·mL-1)购自Solarbio公司;Ex Taq DNA聚合酶、DH5α感受态、DNA胶回收试剂盒和PrimeScriptTM RT reagent kit with gDNA Eraser(Perfect Real Time)购自TaKaRa公司;BbsⅠ-HF限制性内切酶、T4 DNA连接酶购自NEB公司;转染试剂Trans IT-X2TM Dynamic Delivery System购自Mirusbio公司;质粒提取试剂盒QIA prep® Spin Miniprep kit购自Qiagen公司;FastStart Universal SYBR Green MasterBak购自ROCHE公司;DyLight 594 Goat Anti-Mouse IgG和DyLight 488 Goat Anti-Rabbit IgG均购自Abbkine公司。
美国Nano Drop 2000紫外分光光度计;德国耶拿梯度PCR仪;美国ABI公司7500 FAST荧光定量PCR仪;德国Leica公司DMI-6000全自动倒置荧光显微镜。
1.3 gRNA与引物设计根据GenBank数据库中Md5(登录号AF243438.1)和CVI988/Rispens(登录号ABF72323.1)的meq基因序列,使用GenScript’s gRNA在线软件(https://www.genscript.com),分别设计靶向meq 5′端和3′端保守区的gRNA:meq-gRN和meq-gRC,并在5′端添加BbsⅠ酶切位点(表 1)。用Premier Primer 5.0软件设计引物Step2-F和Step2-R用于pX459-gRNA质粒鉴定;meq-F和meq-R用于扩增meq基因,上述引物由生工生物工程(上海)股份有限公司合成。
|
|
表 1 靶向meq基因的gRNA和用于突变体鉴定的引物序列 Table 1 Oligos and primers used for gRNA cloning and PCR identification of mutants |
将gRNA互补双链复性,连接至BbsⅠ-HF酶切回收的pX459质粒载体,转化DH5α感受态,挑单菌落用Step2-F/Step2-R引物对进行PCR,阳性菌送样测序。确认后提取质粒,测定浓度,-20 ℃保存备用。
1.5 质粒转染及病毒感染CEF以1.5×105个·孔-1接种24孔细胞板,置38.5 ℃、5% CO2培养过夜,按照Trans IT-X2转染试剂说明书操作,将pX459-gRNA质粒共转染CEF,继续培养24 h后接种CVI988/Rispens(5 000 PFU·孔-1),置38.5 ℃、5% CO2培养,48 h后取样进行PCR鉴定。
1.6 基因编辑分析胰酶消化“1.5”中CEF转染/感染细胞,取一半样品离心收集细胞,用1×DNA提取缓冲液(10 mmol·L-1 Tris-HCl,1 mmol·L-1 EDTA,25 mmol·L-1 NaCl和200 μg·mL-1蛋白酶K)混悬,65 ℃ 30 min;95 ℃ 5 min,制备总DNA。用meq-F/meq-R引物对进行PCR,反应程序:94 ℃ 3 min;94 ℃ 30 s,58 ℃ 30 s,72 ℃ 1.5 min;30个循环后,72 ℃ 5 min。PCR产物经1.5%琼脂糖凝胶电泳分析,切胶回收目的片段,16 ℃连接至pMD19-T载体,转化DH5α感受态细胞,挑取单菌落用meq-F/meq-R引物对进行PCR鉴定,阳性菌送样测序。
1.7 病毒克隆与纯化取“1.6”中基因编辑阳性的另一半病毒感染细胞,有限稀释后转接6孔板CEF单层,置38.5 ℃、5% CO2培养3~4 d,显微镜下挑取单噬斑,再次转接24孔CEF中,48 h后按“1.6”所述再次PCR,阳性孔同上进行第二轮单噬斑纯化。经过2轮纯化、PCR鉴定均为阳性编辑的病毒噬斑,再次进行测序分析,同时扩大培养,保种冻存。同时,将编辑毒株连续传代,分别收集第5、10、15代病毒样品进行PCR分析。
1.8 IFA鉴定将CVI988/Rispens和meq基因编辑毒株分别接种48孔细胞培养板上的CEF单层,38.5 ℃、5% CO2培养3 d,用预冷的甲醇/丙酮(1:1)溶液固定,200 μL·孔-1,10 min;PBST(含0.05% Tween-20的PBS)洗3遍,加入含5%脱脂奶的PBST封闭,500 μL·孔-1;PBST洗3遍,加入1:2 000稀释的MDV-gB单抗孵育, 50 μL·孔-1, 37 ℃ 30 min;PBST洗3遍,DyLight 594 Goat Anti-Mouse IgG(1:1 000)稀释液孵育, 50 μL·孔-1, 37 ℃ 30 min;PBST洗3遍,再按上述方法加入MDV-MEQ多抗(1:5 000)和DyLight 488 Goat Anti-Rabbit IgG(1:1 000)稀释液;PBST洗3遍后,荧光显微镜下观察,并拍照。
2 结果 2.1 pX459-gRNA质粒构建及鉴定序列比对分析显示:Md5 meq基因全长1 020 bp,CVI988/Rispens meq基因全长1 196 bp。后者在Md5 meq基因575位有1个176 bp的片段插入(图 1),其余序列完全相同。设计2个gRNA meq-gRN和meq-gRC,分别靶向meq基因5′和3′末端保守区域,克隆至pX459质粒并转化DH5α感受态,挑取单菌落用Step-2 F/R引物对进行PCR,结果表明成功构建Cas9/gRNA表达质粒pX459-meq-gRN和pX459-meq-gRC。
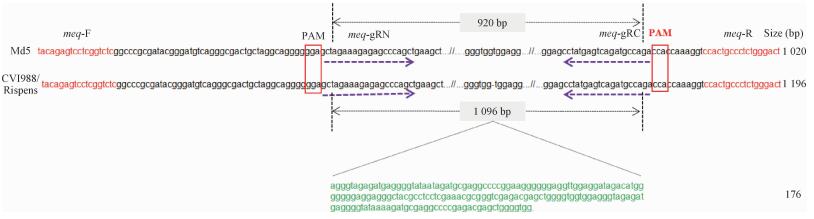
|
图 1 Meq基因座位及gRNA靶点示意图 Fig. 1 Schematics demonstrating the targeting sites of gRNAs used for meq gene editing |
将pX459-meq-gRN和pX459-meq-gRC共转染CEF单层,然后接种CVI988/Rispens,PCR分析结果显示:质粒未转染/病毒感染细胞中仅扩增出1 196 bp的meq特异性条带,而质粒转染/病毒感染细胞中除扩增出1 196 bp meq条带外,还扩增出约100 bp的预期编辑产物条带(图 2A)。小条带产物的测序结果显示,gRN/gRC组合对meq基因进行了有效编辑,双链裂口(DSB)精确发生于2个gRNA PAM序列内侧2~3个碱基处(图 2B、C),产生的基因突变与预期相符。

|
A. meq基因编辑的PCR鉴定;B.野生型和突变型meq基因序列比对;C.突变型meq基因序列图谱; A. PCR analysis of the edited meq genes in CVI988/Rispens viruses; B. Alignment of the wild and mutated types of meq genes; C. Sequence scheme of mutated meq genes; A′. PCR analysis of the stability of meq-deletion in passaged CVI988Δmeq viruses. M.DL2000 DNA marker 图 2 meq基因编辑的PCR鉴定及测序分析 Fig. 2 PCR analysis and DNA sequencing of the gene editing of meq genes |
将上述发生meq基因编辑的感染细胞样品,以有限稀释法转接CEF单层,进行单噬斑纯化和PCR鉴定。结果显示,72个随机挑选的病毒噬斑中,克隆号为C7的病毒噬斑PCR产物仅有100 bp编辑条带,其测序结果和“2.2”测序分析结果完全相同,表明C7为meq基因缺失毒株。对其进行两轮单噬斑纯化,命名为CVI988△meq-C7。连续传代15次,分别收集其第1、5、10、15代次的病毒样品进行PCR分析,结果显示,不同代次病毒样品中均只能扩增出100 bp突变型meq基因产物(图 2A′),表明meq基因缺失非常稳定,连续传代过程中未发生回复突变。
2.4 meq基因缺失的IFA鉴定将CVI988/Rispens或CVI988△meq-C7感染的CEF固定,分别孵育MDV-gB单抗或MDV-MEQ多抗进行IFA染色。结果显示(图 3),用gB单抗孵育的两种病毒感染CEF均呈现红色荧光染色、形态相似的病毒噬斑,表明gB蛋白在两株病毒中均正常表达。用MEQ多抗孵育染色时,绿色荧光病毒噬斑仅出现在CVI988/Rispens感染细胞中,而在CVI988△meq-C7感染细胞中未观察到荧光染色。上述结果证实CVI988△meq-C7的meq基因被编辑缺失,不再表达MEQ蛋白。

|
图 3 病毒感染CEF中gB和MEQ蛋白的IFA分析(比例尺=100 μm) Fig. 3 Expressions of gB and MEQ proteins in virus-infected CEFs detected by IFA (Scale bar=100 μm) |
20世纪90年代,全球广泛使用CVI988/Rispens防控MD,起到了较好的免疫保护效果[20-21],近年来,MDV毒力进一步增强,CVI988/Rispens免疫鸡群中仍能分离到致病性更强的毒株[22]。CVI988/Rispens是MDV自然减毒株,与强毒株相比具有98%的基因组相似性[23],但与毒力密切相关的原癌基因meq却存在较大的序列差异,如在meq基因编码的反式激活结构域有1个176 bp核苷酸的插入,因此也被称为L-meq[24-25]。这种插入造成反式激活区富含脯氨酸重复序列(proline-rich repeats, PRR)的数量增加,导致L-meq基因产物的反式激活区活性不同,可能是CVI988/Rispens疫苗株丧失致癌性的原因[26]。因此,meq和L-meq及其表达的MEQ蛋白,可作为强、弱毒株的鉴别诊断标识。此前利用CRISPR/Cas9基因编辑技术,已实现了HVT基因编辑[27-28]。利用该技术,作者所在课题组也对MD肿瘤细胞整合的病毒pp38和miRNA基因进行了编辑和缺失[29-30]。本研究中,通过设计合成靶向meq基因两端的gRNA组合,利用双gRNA质粒转染和病毒感染的模式对CVI988/Rispens的meq基因进行了编辑,经PCR筛选、测序分析、病毒纯化、IFA鉴定等一系列试验,最终构建了meq基因缺失的CVI988△meq-C7毒株,该毒株可连续传代,稳定性良好。在后续研究中,将其与此前已构建的Md5△meq基因缺失毒株一起,采取交叉筛选的方式可供鉴定不同毒力MDV MEQ蛋白的抗体,为MD感染与免疫的鉴别诊断研究奠定了重要的基础。
4 结论利用CRISPR/Cas9基因编辑技术,通过双gRNA转染/MDV病毒感染的模式,成功构建了meq基因缺失的CVI988△meq-C7毒株,为后续筛选和鉴定MD疫苗株MEQ蛋白的特异性抗体奠定了基础。
| [1] |
SAIF Y M.禽病学[M].苏敬良, 高福, 索勋, 译. 12版.北京: 中国农业出版社, 2012: 129-151.
SAIF Y M. Diseases of poultry[M]. SU J L, GAO F, SUO X, trans. 12th ed. Beijing: China Agriculture Press, 2012: 129-151. (in Chinese) |
| [2] | DAVISON F, NAIR V. Use of Marek's disease vaccines:could they be driving the virus to increasing virulence?[J]. Expert Rev Vaccines, 2005, 4(1): 77–88. |
| [3] | EBINA H, MISAWA N, KANEMURA Y, et al. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus[J]. Sci Rep, 2013, 3: 2510. DOI: 10.1038/srep02510 |
| [4] | SUENAGA T, KOHYAMA M, HIRAYASU K, et al. Engineering large viral DNA genomes using the CRISPR-Cas9 system[J]. Microbiol Immunol, 2014, 58(9): 513–522. DOI: 10.1111/1348-0421.12180 |
| [5] | BI Y W, SUN L, GAO D D, et al. High-efficiency targeted editing of large viral genomes by RNA-guided nucleases[J]. PLoS Pathog, 2014, 10(5): e1004090. DOI: 10.1371/journal.ppat.1004090 |
| [6] | BIERLE C J, ANDERHOLM K M, WANG J B, et al. Targeted mutagenesis of guinea pig cytomegalovirus using CRISPR/Cas9-mediated gene editing[J]. J Virol, 2016, 90(10): 6989–6998. |
| [7] | XU A T, QIN C, LANG Y, et al. A simple and rapid approach to manipulate pseudorabies virus genome by CRISPR/Cas9 system[J]. Biotechnol Lett, 2015, 37(6): 1265–1272. DOI: 10.1007/s10529-015-1796-2 |
| [8] | YUAN M, ZHANG W S, WANG J, et al. Efficiently editing the vaccinia virus genome by using the CRISPR-Cas9 system[J]. J Virol, 2015, 89(9): 5176–5179. DOI: 10.1128/JVI.00339-15 |
| [9] | YUEN K S, CHAN C P, WONG N H, et al. CRISPR/Cas9-mediated genome editing of Epstein-Barr virus in human cells[J]. J Gen Virol, 2015, 96(3): 626–636. DOI: 10.1099/jgv.0.000012 |
| [10] | LIANG X, SUN L Q, YU T, et al. A CRISPR/Cas9 and Cre/Lox system-based express vaccine development strategy against re-emerging pseudorabies virus[J]. Sci Rep, 2016, 6: 19176. DOI: 10.1038/srep19176 |
| [11] | ZOU Z, HUANG K, WEI Y M, et al. Construction of a highly efficient CRISPR/Cas9-mediated duck enteritis virus-based vaccine against H5N1 avian influenza virus and duck tembusu virus infection[J]. Sci Rep, 2017, 7(1): 1478. DOI: 10.1038/s41598-017-01554-1 |
| [12] | PENG Z Y, OUYANG T, PANG D X, et al. Pseudorabies virus can escape from CRISPR-Cas9-mediated inhibition[J]. Virus Res, 2016, 223: 197–205. DOI: 10.1016/j.virusres.2016.08.001 |
| [13] | TANG Y D, LIU J T, WANG T Y, et al. Live attenuated pseudorabies virus developed using the CRISPR/Cas9 system[J]. Virus Res, 2016, 225: 33–39. DOI: 10.1016/j.virusres.2016.09.004 |
| [14] | PAN Q, WANG J, GAO Y L, et al. The natural large genomic deletion is unrelated to the increased virulence of the novel genotype fowl adenovirus 4 recently emerged in China[J]. Viruses, 2018, 10(9): 494. DOI: 10.3390/v10090494 |
| [15] | BORCA M V, HOLINKA L G, BERGGREN K A, et al. CRISPR-Cas9, a tool to efficiently increase the development of recombinant African swine fever viruses[J]. Sci Rep, 2018, 8(1): 3154. |
| [16] | OSTERRIEDER N, KAMIL J P, SCHUMACHER D, et al. Marek's disease virus:from miasma to model[J]. Nat Rev Microbiol, 2006, 4(4): 283–294. DOI: 10.1038/nrmicro1382 |
| [17] | WITTER R L, SCHAT K. Marek's disease[M]//SAIF Y M. Diseases of Poultry. 11th ed. Ames: Iowa State University Press, 2003: 407-464. |
| [18] | SILVA R F, DUNN J R, CHENG H H, et al. A MEQ-deleted Marek's disease virus cloned as a bacterial artificial chromosome is a highly efficacious vaccine[J]. Avian Dis, 2010, 54(2): 862–869. |
| [19] | LI Y P, SUN A J, SU S, et al. Deletion of the meq gene significantly decreases immunosuppression in chickens caused by pathogenic Marek's disease virus[J]. Virol J, 2011, 8: 2. DOI: 10.1186/1743-422X-8-2 |
| [20] | DE BOER G F, GROENENDAL J E, BOERRIGTER H M, et al. Protective efficacy of Marek's disease virus (MDV) CVI-988 CEF65 clone C against challenge infection with three very virulent MDV strains[J]. Avian Dis, 1986, 30(2): 276–283. |
| [21] | RISPENS B H, VAN VLOTEN H, MASTENBROEK N, et al. Control of Marek's disease in the Netherlands. I. Isolation of an avirulent Marek's disease virus (strain CVI 988) and its use in laboratory vaccination trials[J]. Avian Dis, 1972, 16(1): 108–125. |
| [22] | NAIR V. Evolution of Marek's disease-a paradigm for incessant race between the pathogen and the host[J]. Vet J, 2005, 170(2): 175–183. DOI: 10.1016/j.tvjl.2004.05.009 |
| [23] | SPATZ S J, PETHERBRIDGE L, ZHAO Y G, et al. Comparative full-length sequence analysis of oncogenic and vaccine (Rispens) strains of Marek's disease virus[J]. J Gen Virol, 2007, 88(4): 1080–1096. DOI: 10.1099/vir.0.82600-0 |
| [24] | ROSS N L J. T-cell transformation by Marek's disease virus[J]. Trends Microbiol, 1999, 7(1): 22–29. DOI: 10.1016/S0966-842X(98)01427-9 |
| [25] | SCHAT K A, BUCKMASTER A, ROSS L J N. Partial transcription map of Marek's disease herpesvirus in lytically infected cells and lymphoblastoid cell lines[J]. Int J Cancer, 1989, 44(1): 101–109. |
| [26] | CHANG K S, OHASHI K, ONUMA M. Suppression of transcription activity of the MEQ protein of oncogenic Marek's disease virus serotype 1(MDV1) by L-MEQ of non-oncogenic MDV1[J]. J Vet Med Sci, 2002, 64(12): 1091–1095. DOI: 10.1292/jvms.64.1091 |
| [27] | YAO Y X, BASSETT A, NAIR V. Targeted editing of avian herpesvirus vaccine vector using CRISPR/Cas9 nuclease[J]. Intl J Vaccine Technol, 2016, 1: 1–7. |
| [28] | TANG N, ZHANG Y Y, PEDRERA M, et al. A simple and rapid approach to develop recombinant avian herpesvirus vectored vaccines using CRISPR/Cas9 system[J]. Vaccine, 2018, 36(5): 716–722. DOI: 10.1016/j.vaccine.2017.12.025 |
| [29] | ZHANG Y Y, LUO J, TANG N, et al. Targeted editing of the pp38 gene in Marek's disease virus-transformed cell lines using CRISPR/Cas9 System[J]. Viruses, 2019, 11(5): 391. DOI: 10.3390/v11050391 |
| [30] | ZHANG Y Y, TANG N, LUO J, et al. Marek's disease virus-encoded microRNA 155 ortholog critical for the induction of lymphomas is not essential for the proliferation of transformed cell lines[J]. J Virol, 2019, 93(17): e00713–19. |



