2. 黑龙江省农业科学院, 哈尔滨 150086
2. Heilongjiang Academy of Agricultural Sciences, Harbin 150086, China
核不均一核糖核蛋白(heterogeneous nuclear ribonucleoprotein, hnRNPs)是一个保守的核蛋白超家族,在动植物中广泛分布,目前为止,已发现该家族成员有20多种,命名分别从A1至U[1]。大量的研究表明,hnRNPs家族成员在哺乳动物发育中起着至关重要的作用[2],可参与pre-RNA剪接、mRNA运输、定位及翻译等过程[3-5]。hnRNPUL1是hnRNPs家族新成员。hnRNPUL1可与mRNA结合影响RNA的转运和加工[6-7],也可以通过激活多种信号通路促进DNA损伤修复[8-11],还有研究表明,hnRNPUL1与疾病(如早发型心肌梗死)发生相关[12]。
目前已在多物种中对hnRNPUL1基因进行了研究,人hnRNPUL1基因位于19号染色体上,由18个外显子组成,预计编码856个氨基酸残基;牛hnRNPUL1基因位于18号染色体上,由16个外显子组成,预计编码858个氨基酸残基;小鼠hnRNPUL1基因位于7号染色体上,由17个外显子组成,预计编码859个氨基酸残基;这些物种hnRNPUL1均具有保守结构域SAP、SPRY和AAA。但目前尚未见到有关猪hnRNPUL1基因的报道。为进一步揭示猪hnRNPUL1基因功能,本研究以民猪为试验材料,通过RT-PCR方法克隆了猪hnRNPUL1的全长编码区(complete coding sequence, CDS),利用生物信息学方法分析其序列和结构,确定其组织表达谱和亚细胞定位情况,并对其存在的变异剪接体进行鉴定。
1 材料与方法 1.1 试验动物3头3月龄民猪(体重约15 kg,母猪)由黑龙江省农业科学院畜牧研究所科研基地提供,屠宰后立即采集心、肝、脾、肺、肾、胃、大肠、小肠、背肌、腿肌等10个组织样,于液氮中速冻后,-80 ℃冰箱长期保存。
1.2 引物设计与合成通过分析GenBank数据库提供的猪EST、电子预测序列及人、鼠等动物的相应序列,设计H引物,用于扩增猪hnRNPUL1基因的全长编码区。根据克隆测序结果设计引物M1~4、M5、V1~4,分别用于分析变异剪接体、组织表达谱及亚细胞定位情况(详细信息见表 1)。所有引物均利用Premier 5.0软件自行设计,由北京六合华大基因科技股份有限公司合成。
|
|
表 1 引物信息 Table 1 Primer information |
利用Trizol(Invitrogen, Carlsbad, California, USA)提取各组织总RNA,用琼脂糖凝胶电泳和紫外分光光度计检测总RNA的质量和浓度,稀释成1 μg·μL-1备用。按照Prime Script RT reagent Kit(TaKaRa, Dalian, China)说明书进行反转录。将合成的cDNA置于-20 ℃中保存备用。
1.4 hnRNPUL1基因克隆及序列分析以多种组织混合cDNA为模板,利用引物H(表 1)通过PCR方法扩增猪hnRNPUL1基因编码区。反应体系:2×Taq Master Mix 5 μL(Vazyme, Nanjing, China),上下游引物(10 μmol·L-1)各0.4 μL,模板1 μL,ddH2O 3.2 μL,总计10 μL;反应程序:95 ℃ 5 min,(95 ℃ 30 s,60 ℃ 30 s,72 ℃ 1 min 20 s)×30循环,72 ℃ 7 min,16 ℃ 30 s。目的片段胶回收后,插入pMD-18-T载体,转化至大肠杆菌DH5α菌,对平板上的阳性菌落进行菌液PCR和质粒双酶切鉴定后,送到北京六合华大基因科技股份有限公司测序鉴定。
利用SMART程序(http://smart.embl-heidelberg.de/)对猪hnRNPUL1编码的氨基酸序列和蛋白结构域进行分析;利用ProtCompv 9.0 (http://linux1.softberry.com/berry.phtml?group=programs&subgroup=proloc&topic=protcompan)、NLS_Mapper (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi#opennewwindow)和NetNES (http://www.cbs.dtu.dk/services/NetNES/)软件分析猪hnRNPUL1蛋白亚细胞定位情况。
为了更好地分析猪hnRNPUL1基因的可变剪接现象,利用彼此重叠的4对引物(M1~M4,表 1)通过RT-PCR方法进行可变剪接分析,具体方法同上。
1.5 猪hnRNPUL1基因组织表达谱构建将3头猪的同一种组织cDNA进行等量混合后,作为模板,通过Real-time PCR方法构建组织表达谱。利用2×SYBR Green qPCR Master Mix试剂(Bimake, Houston, TX, USA)在ABI QuantStudio3仪器上进行反应。反应体系:SYBR Green Master Mix 10 μL,上下游引物(10 μmol·L-1)各1 μL,cDNA 1 μL,ddH2O 7 μL,总体系为20 μL;反应程序:95 ℃ 5 min,(95 ℃ 15 s,60 ℃ 45 s)×40个循环,95 ℃ 15 s,60 ℃ 60 s,95 ℃ 15 s。每个样品设置3个重复。以β-actin基因为内参,利用2-ΔΔCt方法进行相对表达量计算。
1.6 猪hnRNPUL1亚细胞定位分析以全长编码区的克隆质粒为模板,利用引物V1、V2、V3、V4(表 1)进行PCR扩增后,扩增产物克隆入pMD18-T载体,再通过BamH I和EcoR I酶切位点转接入pEGFP-N1(TaKaRa, Dalian, China)质粒,测序鉴定。
用含10% FBS的DMEM/F12培养基培养PK15细胞,过夜培养待其细胞密度达到70%~80%时,使用脂质体Lipofectamine 2000(Invitrogen, Carlsbad, California, USA)将重组质粒转染至PK15细胞。设置空载体转染组作为阴性对照。转染24 h后使用DAPI(Vazyme, Nanjing, China)染细胞核,在倒置荧光显微镜下观察融合蛋白在PK15细胞中的定位情况。
2 结果 2.1 猪hnRNPUL1基因克隆及序列分析通过RT-PCR和克隆、测序获得了猪hnRNPUL1的全长编码区序列,长度为2 580 bp(图 1、图 2),预期编码859个氨基酸残基的多肽链,分子量为98.06 ku,等电点为8.26,平均亲水性为-0.986(亲水性)。其与人(NP_008971.2)、鼠(NP_659171.1)、牛(NP_001076935.1)hnRNPUL1蛋白的同源性分别为96.97%、89.56%、95.47%。利用SMART程序预测,发现其存在着SAP、SPRY和AAA结构域,分别位于多肽链的3~37、257~390和426~571 aa处。通过BLAT程序(https://genome.ucsc.edu/cgi-bin/hgBlat)分析,确定其含有15个外显子。利用核定位序列预测软件发现,在多肽链的2~30和498~507 aa处存在2个核定位序列(nuclear location sequence, NLS)。将该序列已经提交到GenBank数据库得到序列登录号:MN399154。

|
M. DNA相对分子质量标准,条带长度分别为5 000、3 000、2 000、1 500、1 000、750、500、250、100 bp;1.扩增产物 M. DL5000 marker, composed of 5 000, 3 000, 2 000, 1 500, 1 000, 750, 500, 250 and 100 bp DNA fragments; 1. Amplified products 图 1 RT-PCR电泳检测结果 Fig. 1 Electrophoresis pattern of RT-PCR |

|
E表示外显子,小写字母为引物序列,斜体加粗表示预测的核定位序列,单下划线表示可变剪接体H-1和H-2共有序列,双下划线表示H-2特有序列,边框表示起始密码子和终止密码子,其中TAG为全长扩增产物H的终止密码子,TAA为可变剪接体H-1和H-2的终止密码子 The E represent the exons, the lowercase letters are the primer sequences, the bold italic are putative NLS, the common sequences of transcript variant H-1 and H-2 are underlined, the sequence unique to H-2 is double underlined. The start and stop codons are boxed, of which TAG is the stop codon of H, while TAA is the stop codon of alternative splicing variant H-1 and H-2 图 2 猪hnRNPUL1核苷酸序列 Fig. 2 Nucleotide sequence of porcine hnRNPUL1 |
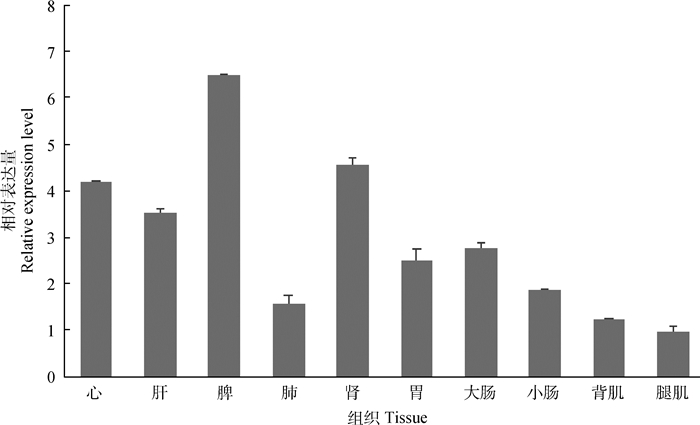
实时荧光定量PCR分析表明,hnRNPUL1基因在所检测的所有组织中均表达,在脾脏中表达量最高,心、肝和肾中的表达量较高,但在肺、胃、大肠、小肠、背肌中表达量较低,在腿肌中表达量最低(图 3)。
|
腿肌组织中的相对表达量作为1,数据为“平均数±标准误” The relative expression level in the leg muscle tissue is used as 1, data is "mean ± standard error" 图 3 猪hnRNPUL1组织表达谱 Fig. 3 Tissue expression profile of porcine hnRNPUL1 gene |
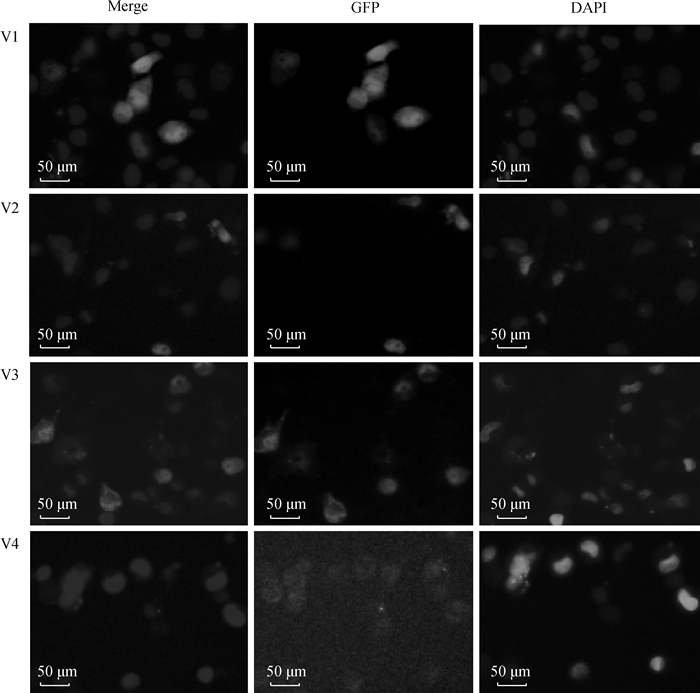
根据亚细胞定位预测结果,首先构建了V1(1~155 aa)和V3(417~738 aa)2个片段的真核表达质粒,转染后发现V1在核质中都有表达,V3仅表达于细胞质中,说明二者不含有NLS。在此基础上,又构建了V2(133~430 aa)和V4(717~859 aa)2个片段的真核表达质粒,发现V2仅表达于细胞核,V4仅表达于细胞质(图 4),说明V2片段存在着NLS。
|
图 4 猪hnRNPUL1基因亚细胞定位 Fig. 4 Subcellular localization of porcine hnRNPUL1 gene |
在克隆测序时,通过菌液PCR对平板上的菌落进行初步鉴定,发现了两个较短的扩增产物,测序后证明是hnRNPUL1基因的可变剪接体,命名为H-1和H-2。与上述克隆的全长CDS相比,二者均缺失外显子2~12的完整序列以及外显子1、13的部分序列;此外,H-2在外显子13末端插入了111 bp(图 2)。H-1和H-2都存在着编码提前终止现象且终止密码子相同,预期编码多肽链长分别为128和166个氨基酸残基,其中前70 aa完全一致,在多肽链的3~37 aa处存在着经典结构域SAP。
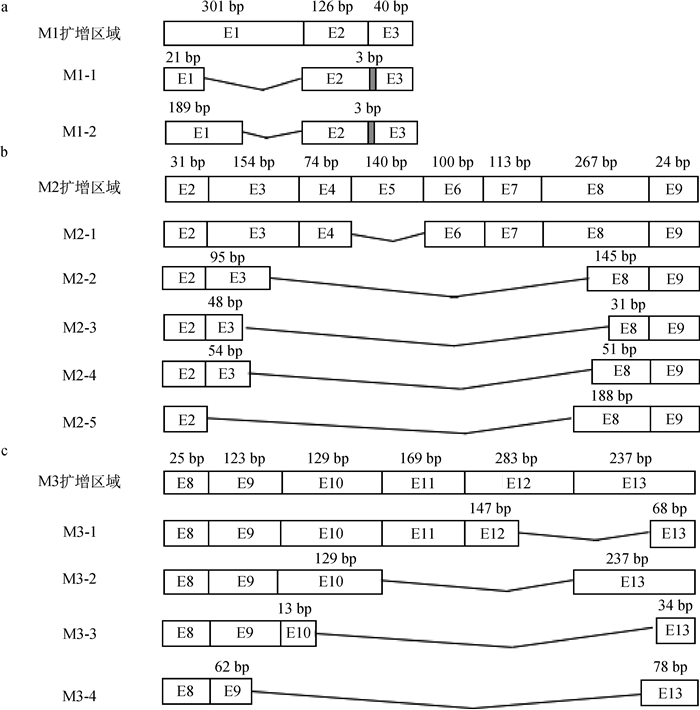
为了更好地分析猪hnRNPUL1基因的可变剪接现象,利用彼此重叠的4对引物(M1~4)通过RT-PCR方法进行可变剪接分析。除M4引物外,M1~M3引物均成功扩增得到了可变剪接片段,3对引物分别获得了2、5和4个可变剪接片段(图 5)。与全长CDS的相应片段比对发现,新发现的可变剪接片段都存在着外显子缺失,剪接方式有外显子跳跃、可变5′剪接位点和可变3′剪接位点,其中外显子8和13存在着较多的可变3′剪接位点,外显子3存在着较多的可变5′剪接位点。M1引物得到的两个可变剪接片段还存在着内含子滞留现象。
|
a. M1引物扩增结果;b. M2引物扩增结果;c. M3引物扩增结果。E表示外显子,折线表示外显子跳跃,灰色阴影方框表示内含子滞留 a. Amplification results of M1 primer; b. Amplification results of M2 primer; c. Amplification results of M3 primer. E indicate exons, the broken lines represent exon skipping, the closed boxes indicate intron retention 图 5 M1、M2、M3扩增区域变异剪接体结构 Fig. 5 Mutant splice variants structure of M1, M2 and M3 amplifying region |
最近的研究表明,hnRNPUL1与聚ADP核糖聚合酶1(poly ADP-ribose polymerase-1, PARP1)有关的DNA损伤修复密切相关,可通过与PARP1的相互作用进而调节PARP1表达的转录调控[13],而PARP1是维持基因组稳定所必需的重要蛋白质之一[14-16]。DNA损伤后不能有效修复会导致损伤积聚和基因组稳定性下降,进而会引起相应遗传性疾病和散发性癌症的发展[17-19]。因此,hnRNPUL1在维持动物机体基因组完整性中发挥作用。
本研究成功克隆了猪hnRNPUL1基因全长CDS序列,发现其在组织中广泛表达,并通过生物信息学分析确定了其编码的多肽链含有hnRNPs家族的经典结构域——SAP、SPRY和AAA。SAP基序通常存在于多种参与转录、DNA修复、RNA处理或凋亡染色质降解的核蛋白中[20-21];具有SPRY结构域的蛋白多参与RNA代谢、细胞内钙释放、调控和发育过程,可能还参与细胞因子信号调控[22-23];AAA基序则分布广泛,参与各种细胞过程,包括膜融合、蛋白水解和DNA复制等[24]。因此,推测猪hnRNPUL1基因同样参与以上多种生理生化进程,对维持动物机体正常的理化反应及功能具有重要意义。
亚细胞定位是分子细胞生物学和蛋白组学一个重要的研究课题,目前多使用绿色荧光蛋白GFP构建融合载体来实现目标蛋白的定位观察[25-28]。通过网站预测发现,该基因具有两个核定位信号,分别位于2~30、498~507 aa处,对应编码区的6~90和1 494~1 521 bp。为鉴定核定位信号位置,首先构建了V1(1~155 aa)和V3(417~738 aa)2个片段的真核表达质粒。发现V1融合蛋白在细胞核与细胞质中均有表达,V3融合蛋白在细胞质中表达。表明这两个区域不存在核定位序列,软件预测结果存在着偏差。作为hnRNPs家族成员,细胞核内定位表达是其发挥功能的基础[29-32]。本研究又继续构建了V2(133~430 aa)和V4(717~859 aa)2个片段的真核表达质粒。结果发现,V2融合蛋白仅在细胞核中表达;V4融合蛋白的表达量比较弱,但能清晰地看到其表达于细胞质中,这可能是由于截短蛋白稳定性降低导致的。
可变剪接现象在真核生物中普遍存在,约有90%人类基因存在可变剪接,是机体在转录后水平调控基因表达的重要手段[33]。研究表明,几乎所有人类基因都存在选择性剪接[34-38]。选择性剪接在控制分化和发育中起着关键作用[39-41],许多重大疾病都与选择性剪接失调有关[42-45]。本试验在鉴定猪hnRNPUL1基因的全长编码区时,获得了2个新的变异剪接体H-1和H-2。为了更好地分析猪hnRNPUL1基因的可变剪接现象,利用彼此重叠的4对引物(M1~M4)通过RT-PCR方法进行鉴定。本试验在其编码区发现了11种新的剪接现象,剪接方式有外显子跳跃、可变5′剪接位点、可变3′剪接位点和内含子滞留。本研究通过菌液PCR结合琼脂糖凝胶电泳检测方法对RT-PCR产物进行筛选,选择长度不同的条带进行测序。由于琼脂糖凝胶电泳的灵敏性较低,无法鉴定长度相同但剪接方式不同的RT-PCR产物,所以不能全面反映猪hnRNPUL1基因的可变剪接情况,但说明了猪hnRNPUL1基因编码区存在着丰富的可变剪接现象。
4 结论通过RT-PCR方法克隆了猪hnRNPUL1基因的全长编码区,确定了其组织表达和亚细胞定位情况,并发现其存在着复杂的可变剪接现象,为研究猪hnRNPUL1基因的生物学功能提供了基础。
| [1] | DREYFUSS G, KIM V N, KATAOKA N. Messenger-RNA-binding proteins and the messages they carry[J]. Nat Rev Mol Cell Biol, 2002, 3(3): 195–205. |
| [2] | ROSHON M J, RULEY H E. Hypomorphic mutation in hnRNP U results in post-implantation lethality[J]. Transgenic Res, 2005, 14(2): 179–192. |
| [3] | GEUENS T, BOUHY D, TIMMERMAN V. The hnRNP family:insights into their role in health and disease[J]. Hum Genet, 2016, 135(8): 851–867. |
| [4] | CARPENTER B, MACKAY C, ALNABULSI A, et al. The roles of heterogeneous nuclear ribonucleoproteins in tumour development and progression[J]. Biochim Biophys Acta, 2006, 1765(2): 85–100. |
| [5] | KRECIC A M, SWANSON M S. hnRNP complexes:composition, structure, and function[J]. Curr Opin Cell Biol, 1999, 11(3): 363–371. |
| [6] | GABLER S, SCHVTT H, GROITL P, et al. E1B 55-kilodalton-associated protein:a cellular protein with RNA-binding activity implicated in nucleocytoplasmic transport of adenovirus and cellular mRNAs[J]. J Virol, 1998, 72(10): 7960–7971. |
| [7] | BACHI A, BRAUN I C, RODRIGUES J P, et al. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates[J]. RNA, 2000, 6(1): 136–158. |
| [8] | BARRAL P M, RUSCH A, TURNELL A S, et al. The interaction of the hnRNP family member E1B-AP5 with p53[J]. FEBS Lett, 2005, 579(13): 2752–2758. |
| [9] | POLO S E, BLACKFORD A N, CHAPMAN J R, et al. Regulation of DNA-end resection by hnRNPU-like proteins promotes DNA double-strand break signaling and repair[J]. Mol Cell, 2012, 45(4): 505–516. |
| [10] | SCHREIBER V, DANTZER F, AME J C, et al. Poly (ADP ribose):novel functions for an old molecule[J]. Nat Rev Mol Cell Biol, 2006, 7(7): 517–528. |
| [11] | SHIFFMAN D, ROWLAND C M, LOUIE J Z, et al. Gene variants of VAMP8 and HNRPUL1 are associated with early-onset myocardial infarction[J]. Arterioscler Thromb Vasc Biol, 2006, 26(7): 1613–1618. |
| [12] | CHATTERJEE N, WALKER G C. Mechanisms of DNA damage, repair, and mutagenesis[J]. Environ Mol Mutagen, 2017, 58(5): 235–263. |
| [13] | HONG Z H, JIANG J, MA J L, et al. The role of hnRPUL1 involved in DNA damage response is related to PARP1[J]. PLoS One, 2013, 8(4): e60208. |
| [14] | JI Y X, WANG Q, ZHAO Q, et al. Autophagy suppression enhances DNA damage and cell death upon treatment with PARP inhibitor Niraparib in laryngeal squamous cell carcinoma[J]. Appl Microbiol Biotechnol, 2019, 103(23-24): 9557–9568. |
| [15] | MACCIÒ A, MADEDDU C. The mechanism of cancer cell death by PARP inhibitors goes beyond DNA damage alone[J]. Int J Cancer, 2019, 145(9): 2594–2596. |
| [16] | CHABANON R M, SORIA J C, LORD C J, et al. Beyond DNA repair:the novel immunological potential of PARP inhibitors[J]. Mol Cell Oncol, 2019, 6(2): 1585170. |
| [17] | VISCONTI R, GRIECO D. New insights on oxidative stress in cancer[J]. Curr Opin Drug Discov Devel, 2009, 12(2): 240–245. |
| [18] | REUTER S, GUPTA S C, CHATURVEDI M M, et al. Oxidative stress, inflammation, and cancer:how are they linked?[J]. Free Radic Biol Med, 2010, 49(11): 1603–1616. |
| [19] | SHARMA A, JOHNSON A. Exosome DNA:critical regulator of tumor immunity and a diagnostic biomarker[J]. J Cell Physiol, 2020, 235(3): 1921–1932. |
| [20] | NAYLER O, STRÄTLING W, BOURQUIN J P, et al. SAF-B protein couples transcription and pre-mRNA splicing to SAR/MAR elements[J]. Nucleic Acids Res, 1998, 26(15): 3542–3549. |
| [21] | ARAVIND L, KOONIN E V. SAP-a putative DNA-binding motif involved in chromosomal organization[J]. Trends Biochem Sci, 2000, 25(3): 112–114. |
| [22] | YAP M W, NISOLE S, STOYE J P. A single amino acid change in the SPRY domain of human Trim5α leads to HIV-1 restriction[J]. Curr Biol, 2005, 15(1): 73–78. |
| [23] | RHODES D A, DE BONO B, TROWSDALE J. Relationship between SPRY and B30.2 protein domains.Evolution of a component of immune defence?[J]. Immunology, 2005, 116(4): 411–417. |
| [24] | OGURA T, WILKINSON A J. AAA+ superfamily ATPases:common structure--diverse function[J]. Genes Cells, 2001, 6(7): 575–597. |
| [25] | MISTELI T, SPECTOR D L. Applications of the green fluorescent protein in cell biology and biotechnology[J]. Nat Biotechnol, 1997, 15(10): 961–964. |
| [26] |
张霞, 宋雪, 王配, 等. 版纳微型猪近交系CMAH基因克隆及亚细胞定位研究[J]. 河北农业大学学报, 2019, 42(3): 94–98.
ZHANG X, SONG X, WANG P, et al. Cloning and subcellular location of CMAH gene in Banna mini-pig inbred line[J]. Journal of Hebei Agricultural University, 2019, 42(3): 94–98. (in Chinese) |
| [27] |
范兴兴, 李新建, 李改英, 等. 猪Fosl2基因克隆、结构预测及其mRNA发育性表达变化[J]. 畜牧兽医学报, 2015, 46(2): 211–218.
FAN X X, LI X J, LI G Y, et al. Gene cloning, bioinformatics analysis and developmental expression of Fosl2 gene in pig[J]. Acta Veterinaria et Zootechnica Sinica, 2015, 46(2): 211–218. (in Chinese) |
| [28] |
赵为民, 涂枫, 方晓敏, 等. 猪PKM2基因的序列分析与组织表达及亚细胞定位[J]. 湖南农业大学学报:自然科学版, 2019, 45(1): 68–77.
ZHAO W M, TU F, FANG X M, et al. Sequence analysis and tissue expression and subcellular localization of porcine PKM2 gene[J]. Journal of Hunan Agricultural University:Natural Sciences, 2019, 45(1): 68–77. (in Chinese) |
| [29] | CAPPELLI S, ROMANO M, BURATTI E. Systematic analysis of gene expression profiles controlled by hnRNP Q and hnRNP R, two closely related human RNA binding proteins implicated in mRNA processing mechanisms[J]. Front Mol Biosci, 2018, 5: 79. |
| [30] | GASPERINI L, ROSSI A, CORNELLA N, et al. The hnRNP RALY regulates PRMT1 expression and interacts with the ALS-linked protein FUS:implication for reciprocal cellular localization[J]. Mol Biol Cell, 2018, 29(26): 3067–3081. |
| [31] | RUAN Q T, YAZDANI N, BLUM B C, et al. A mutation in Hnrnph1 that decreases methamphetamine-induced reinforcement, reward, and dopamine release and increases synaptosomal hnRNP H and mitochondrial proteins[J]. J Neurosci, 2019, 11: 1808–1819. |
| [32] | IZUMI H, FUNA K. Telomere function and the G-Quadruplex formation are regulated by hnRNP U[J]. Cells, 2019, 8(5): 390. |
| [33] | NAGASAKI H, ARITA M, NISHIZAWA T, et al. Automated classification of alternative splicing and transcriptional initiation and construction of visual database of classified patterns[J]. Bioinformatics, 2006, 22(10): 1211–1216. |
| [34] | PAN Q, SHAI O, LEE L J, et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing[J]. Nat Genet, 2008, 40(12): 1413–1415. |
| [35] |
邵红伟, 张文峰, 胡青莲, 等. 两种存活蛋白变异剪接体在HeLa细胞中的表达定位及相互作用[J]. 中国生物化学与分子生物学报, 2009, 25(12): 1131–1136.
SHAO H W, ZHANG W F, HU Q L, et al. Expression and location of two survivin splice variants and their interaction in HeLa Cells[J]. Chinese Journal of Biochemistry and Molecular Biology, 2009, 25(12): 1131–1136. (in Chinese) |
| [36] |
赵晋枫, 武宝爱. 有氧运动影响低密度脂蛋白受体pre-mRNA的选择性剪接及组蛋白修饰机制研究[J]. 山东体育学院学报, 2018, 34(6): 78–83.
ZHAO J F, WU B A. Histone modifications involved in alternative splicing of LDLR pre-mRNA and the influence of aerobic exercise[J]. Journal of Shandong Sport University, 2018, 34(6): 78–83. (in Chinese) |
| [37] |
张天, 李祥龙, 周荣艳, 等. 不同毛色山羊皮肤组织Agouti基因剪接体类型研究[J]. 畜牧兽医学报, 2015, 46(11): 1934–1943.
ZHANG T, LI X L, ZHOU R Y, et al. Study on Agouti spliceosome types in goat skin with different coat color[J]. Acta Veterinaria et Zootechnica Sinica, 2015, 46(11): 1934–1943. (in Chinese) |
| [38] |
李利族, 韩丽鑫, 王金奎, 等. 猪PILRA基因克隆及变异剪接体鉴定[J]. 遗传, 2015, 37(9): 926–931.
LI L Z, HAN L X, WANG J K, et al. Cloning and identification of splice variants of the porcine PILRA gene[J]. Hereditas (Beijing), 2015, 37(9): 926–931. (in Chinese) |
| [39] | NUTTER C A, BUBENIK J L, OLIVEIRA R, et al. Cell-type-specific dysregulation of RNA alternative splicing in short tandem repeat mouse knockin models of myotonic dystrophy[J]. Genes Dev, 2019, 33(23-24): 1635–1640. |
| [40] |
谢玉霜, 包永芬, 白育庭, 等. 变异剪接体的结构、功能及其在疾病治疗中的应用[J]. 生命的化学, 2019, 39(4): 660–664.
XIE Y S, BAO Y F, BAI Y T, et al. The structure and function of the spliceosome and its application in the diseases therapy[J]. Chemistry of Life, 2019, 39(4): 660–664. (in Chinese) |
| [41] | JIA R, AJIRO M, YU L L, et al. Oncogenic splicing factor SRSF3 regulates ILF3 alternative splicing to promote cancer cell proliferation and transformation[J]. RNA, 2019, 25(5): 630–644. |
| [42] | BRAGGIN J E, BUCKS S A, COURSE M M, et al. Alternative splicing in a presenilin 2 variant associated with Alzheimer disease[J]. Ann Clin Trans Neurol, 2019, 6(4): 762–777. |
| [43] | WU ZE, LIANG J H, WANG C P, et al. Alternative splicing provides a mechanism to regulate LlHSFA3 function in response to heat stress in lily[J]. Plant Physiol, 2019, 181(4): 1651–1667. |
| [44] | LI L P, YAN S X, ZHANG H, et al. Interaction of hnRNP K with MAP 1B-LC1 promotes TGF-β1-mediated epithelial to mesenchymal transition in lung cancer cells[J]. BMC Cancer, 2019, 19(1): 894. |
| [45] | LIANG S Z, YU B, QIAN D M, et al. Human cytomegalovirus ie2 affects the migration of glioblastoma by mediating the different splicing patterns of RON through hnRNP A2B1[J]. Neuroreport, 2019, 30(12): 805–811. |



