2. 国家北方山区农业工程技术研究中心, 保定 071000;
3. 河北省山区农业工程技术研究中心, 保定 071000;
4. 河北工程大学 生命科学与食品工程学院, 邯郸 056038
2. National Engineering Research Center for Agriculture in Northern Mountainous Areas, Baoding 071000, China;
3. Engineering Research Center for Agriculture in Hebei Mountainous Areas, Baoding 071000, China;
4. College of Life Sciences and Food Engineering, Hebei University of Engineering, Handan 056038, China
繁殖力对母猪来说是一个至关重要的经济因素,因而提高猪繁殖力是养猪业需要解决的关键性问题。众多研究显示,类固醇激素合成能力的强弱是影响繁殖力的一个重要因素[1]。近年来研究发现,类固醇合成的快速调节机制对于调节动物的生殖活动必不可少,卵泡发育与类固醇激素的合成与分泌密切相关。类固醇合成快速调节蛋白(steroidogenic acute regulatory protein, StAR)作为胆固醇由线粒体外膜向内膜转运的桥梁,参与了类固醇激素的合成,该蛋白由StAR基因所编码。在雌性动物卵巢中,排卵、卵泡的生长与闭锁,以及成熟卵泡内细胞的分化等一系列生理过程均受到类固醇激素的调控。相关研究显示,排卵前卵泡分泌的主要类固醇激素是雌激素,在排卵后,低密度脂蛋白通过上调类固醇合成关键基因StAR和P450的表达量来调控其孕酮合成水平[2],并促使卵泡由分泌雌激素转变为大量分泌孕酮[1, 3],若降低StAR基因的表达水平则可致使卵巢孕酮合成水平下降,此时配种后受胎率明显下降,可见在哺乳动物中,StAR基因是调控类固醇激素合成的一个关键因素,然而该基因的表达也受到多种因素的影响,如胰岛素、LH、FSH等激素[4-6]和IGF1、TGF-β多种生长因子[7]以及转录因子[8]等多种方式。此外,在表观遗传修饰方面相关研究还发现,大鼠颗粒细胞向黄体细胞分化过程中StAR基因启动子区会发生组蛋白修饰[9-10];很多转录因子如cAMP response element-binding protein (CREB)、GATA4、CCAAT/enhancer binding proteins(C/EBPs)参与了大鼠StAR启动子的调控[11-12]。然而,猪StAR基因的调控机制目前尚未见报道。
为了解猪StAR基因表达的调控机制,本研究分析了猪StAR基因5′非翻译区和启动子区,测定StAR基因的5′侧翼区序列的转录活性,筛选出该基因启动子转录调控区域,探索其可能的调控机制,为进一步揭示猪StAR基因的转录调控机制提供理论基础。
1 材料与方法 1.1 材料人肾上皮细胞系(293T)由本实验室保存;pGL3-Basic、pRL-TK载体、T4DNA Ligase、双荧光素酶报告基因检测试剂盒均购自Promega公司;TailGen DNA Kit购自康为世纪公司;SanPrep柱式DNA胶回收试剂盒、SanPrep柱式质粒小量抽提试剂盒购自生工生物工程(上海)股份有限公司;无内毒素质粒大提试剂盒购自北京天根公司;转染试剂LipofectamineTM 2000购自IVGN公司;限制性内切酶KpnⅠ和Hind Ⅲ均购自TaKaRa公司;Trans5α感受态细胞、Trans Taq-T DNA聚合酶、TransSerumTM HQ Fetal Bovine Serum等均购自于北京TransGen生物技术有限公司;Hyclone DMEM/ HIGH GLUCOSE培养基购自Thermo公司;DL5000 DNA Marker购自北京博奥龙免疫技术有限公司;0.25%胰蛋白酶(Trypsin-EDTA)、Opti-MEM培养基购自Gibco公司;引物合成和测序均由北京华大科技公司完成。
1.2 方法 1.2.1 猪StAR基因5′侧翼区序列分析及引物设计检索并下载猪StAR基因(Ensembl数据库:ENSSSCG00000034942)的参考序列,通过基因启动子在线预测分析软件MethPrimer (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi)预测CpG岛;通过在线软件:AliBaba2.1 (http://www.gene-regulation.com/pub/programs.html)、JASPAR数据库(http://jaspar.binf.ku.dk/)预测转录因子结合位点。根据pGL3-Basic质粒的多克隆位点信息,以StAR基因5′ UTR上游序列约2 000 bp为模板,利用Primer Premier 5.0软件设计扩增包含StAR基因启动子区片段的特异引物,分别在上、下游引物5′端引入限制性内切酶KpnⅠ和Hind Ⅲ的酶切位点及其对应的保护碱基,构建载体片段的引物序列详见表 1。
|
|
表 1 本研究所用引物信息 Table 1 The informations of primer used in this study |
提取大白猪耳组织基因组DNA,以基因组DNA为模板,采用PCR方法,利用表 1中引物扩增猪StAR基因启动子区片段,PCR反应体系:10×buffer 2.5 μL,Trans Taq-T DNA聚合酶(5 U·μL-1) 0.7 μL,2.5 mmol·L-1 dNTPs 2.0 μL,上、下游引物(10 pmol·μL-1)各1.0 μL,各模板DNA 1.0 μL,加ddH2O至25.0 μL;PCR反应条件:94 ℃预变性5 min;94 ℃变性30 s,65 ℃退火30 s,72 ℃延伸2 min,34个循环;72 ℃延伸10 min。StAR-1~StAR-4为首次缺失片段,StAR-5~StAR-10为二次缺失片段。
1.2.3 pGL3-StAR启动子不同长度片段重组载体的构建将测序正确的上述10个PCR产物及其pGL3-Basic质粒分别利用限制性内切酶KpnⅠ和Hind Ⅲ进行双酶切,凝胶回收酶切产物后,采用T4连接酶分别将10个片段与pGL3-Basic连接,连接产物转化至感受态细胞,经菌液PCR鉴定,阳性克隆者测序,并进行质粒双酶切,分别命名为pGL3-1939/-103、pGL3-1311/-103、pGL3-470/-103、pGL3-196/-103、pGL3-1939/+127、pGL3-1311/+127、pGL3-470/+127、pGL3-196/+127、pGL3-41/+127、pGL3-120/-17。
1.2.4 细胞培养、转染及活性检测将293T细胞接种于含10%胎牛血清的DMEM培养基中,在含5% CO2的37 ℃培养箱中进行培养。在转染前1 d,将生长状态良好的细胞接种于不含抗生素的24孔板中,待细胞融合度达80%~90%时,分别将重组报告基因质粒、内参质粒pRL-TK与转染试剂LipofectamineTM 2000加入至Opti-MEM培养基中,并将其混合物转染至293T细胞,培养48 h后裂解细胞,利用双荧光素酶报告基因检测试剂盒,采用化学发光仪(Promega)测定荧光素酶活性,试验重复3次及以上,每次3个平行。
1.2.5 统计分析对各组试验至少重复3次,利用单因素方差分析检验不同片段酶活性的差异性,Duncan′s检验法进行多重比较,数据均采用“平均值±标准差”表示。数据利用SPSS 19.0统计软件进行分析。
2 结果 2.1 StAR基因5′侧翼区序列分析利用启动子分析软件对猪StAR基因5′侧翼区序列特征信号进行了分析,结果显示,该序列不具有真核生物启动子典型的TATA-box,此外在该启动子区域还发现可能存在GATA2、GATA4、SP1、ZNF263、Hoxa9、KLF16、ZNF740等多种转录因子结合位点。一般认为CpG岛大于100 bp,且GC含量大于50%[13],本试验利用MethPrimer软件预测发现,目的基因StAR启动子不存在CpG岛(图 1)。
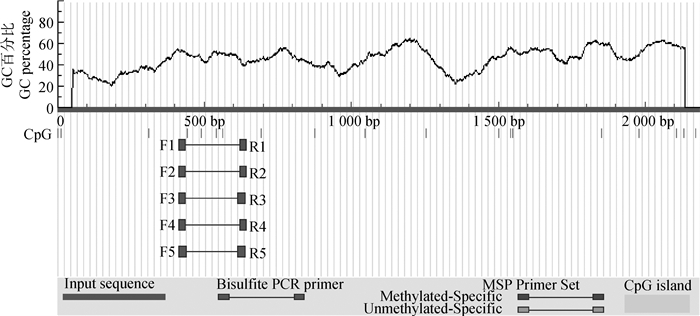
|
F1~F5.亚硫酸盐PCR上游引物;R1~R5.亚硫酸盐PCR下游引物 F1-F5. Bisulfite PCR left primers; R1-R5. Bisulfite PCR right primers 图 1 CpG岛预测结果 Figure 1 CpG island prediction results |
通过PCR扩增得到猪StAR基因启动子5′端10个不同片段,PCR产物分别经1%琼脂糖凝胶电泳检测。如图 2所示,扩增出的不同长度片段与预期片段大小相一致(图 2)。
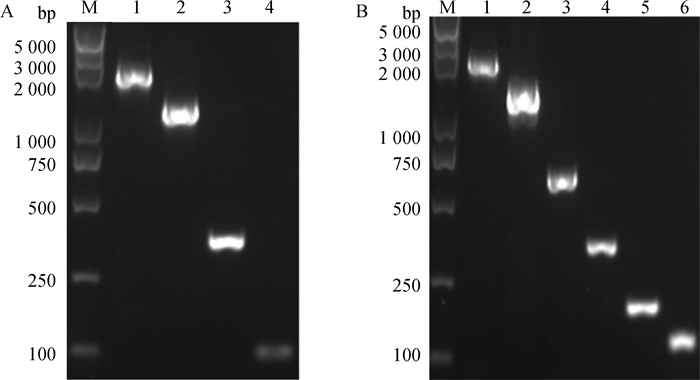
|
M. DNA相对分子质量标准; 1~4. pGL3-1~pGL3-4不同长度扩增片段(A);1~6. pGL3-5~pGL3-10不同长度扩增片段(B) M. DL5000 DNA marker; 1-4. Different length of amplified fragments of pGL3-1-pGL3-4(A); 1-6. Different length of amplified fragments of pGL3-5-pGL3-10(B) 图 2 猪StAR基因启动子不同长度片段PCR扩增产物凝胶电泳图 Figure 2 Agarose gel electropherogram of PCR products of the fragments with different length in swine StAR promoter |
分别将StAR基因启动子系列片段(pGL3-1~pGL3-10)和pGL3-Basic进行双酶切,酶切产物经凝胶回收后构建不同长度的StAR基因启动子重组质粒,经菌液PCR鉴定,为阳性克隆者进行测序和质粒双酶切。重组质粒双酶切结果显示,各重组质粒均产生pGL3-Basic载体片段及各自相应的片段(图 3);测序结果与数据库中猪的StAR基因5′端序列相一致。因而,猪StAR基因启动子缺失片段的荧光素酶报告基因载体构建成功。
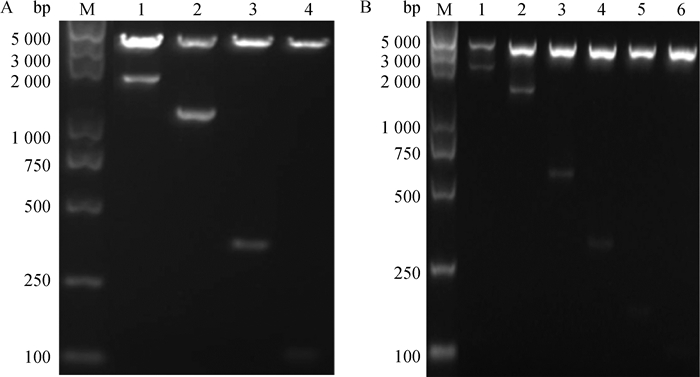
|
M. DNA相对分子质量标准; 1~4. pGL3-1~pGL3-4片段酶切鉴定(A);1~6. pGL3-5~pGL3-10片段酶切鉴定(B) M. DL5000 DNA marker; 1-4. Enzyme digestion of pGL3-1-pGL3-4 fragments(A); 1-6. Enzyme digestion of pGL3-5-pGL3-10 fragments(B) 图 3 pGL3-StAR重组质粒酶切产物凝胶电泳图 Figure 3 Agarose gel electropherogram of pGL3-StAR by enzyme digestion |
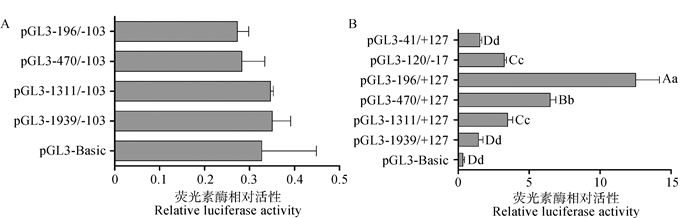
将pEGFPN1质粒转染至293T细胞,转染6 h后观察细胞生长状态良好,继续培养至48 h后,荧光显微镜下检测转染效率,如图 4所示,经过计算超过70%以上的细胞中可以观察到绿色荧光信号。以pGL3-Basic质粒作为阴性对照,将上述所构建的4个重组质粒分别与pRL-TK共转染至293T细胞,继续培养48 h后利用双荧光素酶报告基因检测试剂盒测定萤火虫荧光素酶和内对照海肾荧光素酶荧光值,由图 5A可知,所构建重组质粒pGL3-1939/-103、pGL3-1311/-103、pGL3-470/-103、pGL3-196/-103的荧光素酶活性与pGL3-Basic对照组相比均差异不显著(P>0.05),说明猪StAR基因-1939~-103 bp(以转录起始位点为+1)区域不具有转录活性。
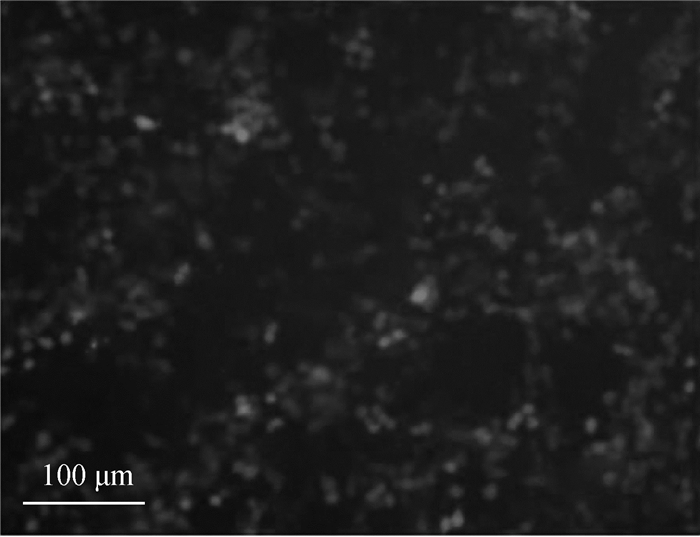
|
图 4 转染后的293T细胞 Figure 4 The 293T cells after transfection |
|
各重组质粒荧光素酶活性相比,相同小写字母标注者为差异不显著(P>0.05), 不同小写字母标注者为差异显著(P < 0.05),不同大写字母标注者为差异极显著(P < 0.01) Bars with the same small letter mean no significant difference (P>0.05), bars with the different small letters mean significant difference (P < 0.05), bars with different capital letters mean extremely significant difference (P < 0.01) among different luciferase activity of recombinant vectors 图 5 重组质粒pGL3-StAR转染293T细胞的活性分析 Figure 5 Activity analysis of recombinant vectors pGL3-StAR in 293T cells |
进而构建pGL3-1939/+127、pGL3-1311/+127、pGL3-470/+127、pGL3-196/+127并转染293T细胞后检测其荧光值,由图 5B可知,与先前构建的4个片段重组质粒相比,片段-1 311~+127、-470~+127、-196~+127、-120~-17活性极显著增加(P < 0.01),其中片段-196~+127 bp活性值最高,且极显著高于其他缺失片段(P < 0.01),表明-103~+127 bp这一区域存在正调控元件。进一步将-196~+127截短为-41~+127,构建pGL3-41/+127重组质粒并转染细胞后检测其荧光值,结果显示,该区域不存在转录活性,说明-41~-196 bp存在正调控元件,进而构建pGL3-120/-17重组质粒,结果显示,该区域存在转录活性,进一步证明-41~-196 bp这一区域存在着重要的正调控元件。
3 讨论类固醇激素合成急性调节蛋白(StAR)位于相关细胞的线粒体膜上,且具有高度的组织特异性,参与类固醇激素的合成,调节胆固醇从线粒体外膜向线粒体内膜的转运,是肾上腺皮质和生殖腺类固醇生成细胞中合成类固醇激素所需的物质[14]。StAR基因在转录及翻译水平上受多种促激素和细胞因子的调控,进而影响着机体类固醇激素的合成[15-17]。在人类中,StAR基因突变可致使其蛋白失去功能,肾上腺和性腺类固醇合成受阻,从而引起一种常染色体隐性遗传病即先天性脂样肾功能增生综合征;在小鼠上,StAR基因在肾上腺、睾丸、卵巢组织中特异性表达,缺失该基因的小鼠发育会受阻,寿命会缩短[18]。卵巢作为雌性动物重要的生殖器官,其功能主要是产生卵细胞以及分泌类固醇激素。研究者在极端高、低产仔数的约克夏母猪卵巢转录组测序中发现了StAR基因在高产组高表达的现象,经筛选分析将该基因作为猪繁殖力和产仔数相关的候选基因[19]。在猪上,StAR基因的调控机制和信号通路尚未研究清楚。因此,本试验对猪StAR启动子区转录活性进行了研究。
基于生物信息学方法对猪StAR基因5′端序列进行分析,结果发现,猪StAR基因5′端序列不存在典型的顺式元件TATA-box,事实上TATA-box是构成真核生物典型的核心启动子元件之一,但TATA-box也并不总是存在于一个基本的启动子区域[20]。为了研究StAR基因启动子转录活性及其调控机制,本试验采用构建启动子缺失片段的方法[21-22],通过构建荧火虫荧光素酶重组载体,转染至293T细胞,利用双荧光素酶检测系统对构建的不同长度启动子区片段进行活性检测。结果显示,猪StAR基因的启动子区域-196~-127 bp活性值最高。采用JASPAR的在线程序MatInspector功能来预测潜在的转录因子结合位点,在猪StAR启动子的-196 ~-41 bp区域存在转录因子GATA4、GATA6、SF-1、SP1、ZNF263、ZNF740、ATF4的结合位点。研究报道表明,GATA4在类固醇生成、性别分化等方面具有重要的作用,GATA4和GATA6在肾上腺和性腺中高表达[23]。在哺乳动物卵巢组织中的作用主要是由cAMP通过PKA途径增加GATA4磷酸化水平,进而调节GATA依赖性靶基因的表达,如类固醇合成关键基因StAR和Cyp17等对卵泡发育有重要作用的靶基因[24];研究还发现,StAR基因在猪发情周期的各个阶段也均有表达[4],该基因启动子区GATA4结合区序列缺失会影响FSH诱导的StAR启动子的活性[25-26]。做为GATA家族的另一成员,GATA6不仅可以调节心、肺组织中相关基因的表达,对组织的正常发育也起到了重要作用,而且在生殖腺中被认为是StAR基因的正向调控因子[27-28]。类固醇合成因子-1(SF-1)与StAR基因之间的相互作用已通过体外试验证明:在尼古丁处理的NCI-H295A细胞中,SF-1在StAR基因表达量降低时,类固醇合成水平也随着降低[29],可见转录因子SF-1也是激活StAR的潜在正调控因子。对于ZNF转录因子,目前在植物中只是作为顺式作用元件在代谢物生物合成中起重要作用[30-31];在动物中,基于ZNF与StAR基因的调控关系,将来可以直接采用电泳迁移率变动分析(EMSA)和染色质免疫沉淀等方法来进一步研究ZNF的调节功能。
4 结论本试验通过对StAR基因进行生物信息学分析,构建猪StAR基因启动子区不同长度片段的重组载体,并利用双报告基因报告系统检测其活性,证实了StAR基因的5′侧翼区序列具有启动子活性。初步确定了猪StAR基因的启动子区域,找到了该基因启动子的核心区域和主要调控区域,为进一步研究猪StAR基因转录调控机制提供理论依据。
| [1] |
张金友. 溶酶体参与LDL促牛颗粒细胞StAR基因表达和孕酮合成的研究[D]. 哈尔滨: 东北农业大学, 2015.
ZHANG J Y. Lysosomes are involved in induction of steroidogenic acute regulatory protein (StAR) gene expression and progesterone synthesis through low density lipoprotein in cultured bovine granulosa cells[D]. Harbin: Northeast Agricultural University, 2015. (in Chinese) |
| [2] | REYLAND M E, EVANS R M, WHITE E K. Lipoproteins regulate expression of the steroidogenic acute regulatory protein (StAR) in mouse adrenocortical cells[J]. J Biol Chem, 2000, 275(47): 36637–36644. DOI: 10.1074/jbc.M006456200 |
| [3] | MAMLUK R, GREBER Y, MEIDAN R. Hormonal regulation of messenger ribonucleic acid expression for steroidogenic factor-1, steroidogenic acute regulatory protein, and cytochrome P450 side-chain cleavage in bovine luteal cells[J]. Biol Reprod, 1999, 60(3): 628–634. DOI: 10.1095/biolreprod60.3.628 |
| [4] | SEKAR N, LAVOIE H A, VELDHUIS J D. Concerted regulation of steroidogenic acute regulatory gene expression by luteinizing hormone and insulin (or insulin-like growth factor Ⅰ) in primary cultures of porcine granulosa-luteal cells[J]. Endocrinology, 2000, 141(11): 3983–3992. DOI: 10.1210/endo.141.11.7763 |
| [5] | SEKAR N, GARMEY J C, VELDHUIS J D. Mechanisms underlying the steroidogenic synergy of insulin and luteinizing hormone in porcine granulosa cells:Joint amplification of pivotal sterol-regulatory genes encoding the low-density lipoprotein (LDL) receptor, steroidogenic acute regulatory (StAR) protein and cytochrome P450 side-chain cleavage (P450scc) enzyme[J]. Mol Cell Endocrinol, 2000, 159(1-2): 25–35. DOI: 10.1016/S0303-7207(99)00203-8 |
| [6] | LUO W X, GUMEN A, HAUGHIAN J M, et al. The role of luteinizing hormone in regulating gene expression during selection of a dominant follicle in cattle[J]. Biol Reprod, 2011, 84(2): 369–378. DOI: 10.1095/biolreprod.110.085274 |
| [7] | FANG L L, CHANG H M, CHENG J C, et al. TGF-β downregulates StAR expression and decreases progesterone production through Smad3 and ERK1/2 signaling pathways in human granulosa cells[J]. J Clin Endocrinol Metab, 2014, 99(11): E2234–E2243. DOI: 10.1210/jc.2014-1930 |
| [8] | HUI Y Y, LAVOIE H A. GATA4 reduction enhances 3', 5'-cyclic adenosine 5'-monophosphate-stimulated steroidogenic acute regulatory protein messenger ribonucleic acid and progesterone production in luteinized porcine granulosa cells[J]. Endocrinology, 2008, 149(11): 5557–5567. DOI: 10.1210/en.2008-0484 |
| [9] | YAMASHITA H, MURAYAMA C, TAKASUGI R, et al. BMP-4 suppresses progesterone production by inhibiting histone H3 acetylation of StAR in bovine granulosa cells in vitro[J]. Mol Cell Biochem, 2011, 348(1-2): 183–190. DOI: 10.1007/s11010-010-0653-9 |
| [10] | LEE L, ASADA H, KIZUKA F, et al. Changes in histone modification and DNA methylation of the StAR and Cyp19a1 promoter regions in granulosa cells undergoing luteinization during ovulation in rats[J]. Endocrinology, 2013, 154(1): 458–470. DOI: 10.1210/en.2012-1610 |
| [11] | MANNA P R, STOCCO D M. Crosstalk of CREB and Fos/Jun on a single cis-element:Transcriptional repression of the steroidogenic acute regulatory protein gene[J]. J Mol Endocrinol, 2007, 39(4): 261–277. DOI: 10.1677/JME-07-0065 |
| [12] | SILVERMAN E, YIVGI-OHANA N, SHER N, et al. Transcriptional activation of the steroidogenic acute regulatory protein (StAR) gene:GATA-4 and CCAAT/enhancer-binding protein β confer synergistic responsiveness in hormone-treated rat granulosa and HEK293 cell models[J]. Mol Cell Endocrinol, 2006, 252(1-2): 92–101. DOI: 10.1016/j.mce.2006.03.008 |
| [13] | LI L C, DAHIYA R. MethPrimer:Designing primers for methylation PCRs[J]. Bioinformatics, 2002, 18(11): 1427–1431. DOI: 10.1093/bioinformatics/18.11.1427 |
| [14] | MILLER W L, AUCHUS R J. The molecular biology, biochemistry, and physiology of human steroidogenesis and its disorders[J]. Endocr Rev, 2011, 32(1): 81–151. DOI: 10.1210/er.2010-0013 |
| [15] | STRICKLAND J, MCILMOIL S, WILLIAMS B J, et al. Interleukin-6 increases the expression of key proteins associated with steroidogenesis in human NCI-H295R adrenocortical cells[J]. Steroids, 2017, 119: 1–17. DOI: 10.1016/j.steroids.2016.12.014 |
| [16] | BAHAT A, PERLBERG S, MELAMED-BOOK N, et al. Transcriptional activation of LON gene by a new form of mitochondrial stress:A role for the nuclear respiratory factor 2 in StAR overload response (SOR)[J]. Mol Cell Endocrinol, 2015, 408(7): 62–72. |
| [17] | BAHAT A, PERLBERG S, MELAMED-BOOK N, et al. StAR enhances transcription of genes encoding the mitochondrial proteases involved in its own degradation[J]. Mol Endocrinol, 2014, 28(2): 208–224. DOI: 10.1210/me.2013-1275 |
| [18] | ISHⅡ T, HASEGAWA T, PAI C I, et al. The roles of circulating high-density lipoproteins and trophic hormones in the phenotype of knockout mice lacking the steroidogenic acute regulatory protein[J]. Mol Endocrinol, 2002, 16(10): 2297–2309. DOI: 10.1210/me.2001-0320 |
| [19] |
黄龙. 基于卵巢RNA组学鉴定影响猪产仔数性状的候选基因及microRNA[D]. 合肥: 安徽农业大学, 2016.
HUANG L. Identification of candidate genes and microRNAs affecting pig litter size by ovarian RNomics[D]. Hefei: Anhui Agricultural University, 2016. (in Chinese) |
| [20] | GOODRICH J A, TJIAN R. Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation[J]. Nat Rev Genet, 2010, 11(8): 549–558. DOI: 10.1038/nrg2847 |
| [21] | ZHAO X, MO D L, LI A N, et al. Characterization and transcriptional regulation analysis of the porcine PAQR6 gene[J]. DNA Cell Biol, 2011, 30(11): 947–954. DOI: 10.1089/dna.2011.1262 |
| [22] | TAO H, MEI S Q, ZHANG X Y, et al. Transcription factor C/EBPβ and 17β-estradiol promote transcription of the porcine p53 gene[J]. Int J Biochem Cell Biol, 2014, 47(2): 76–82. |
| [23] | KETOLA I, RAHMAN N, TOPPARI J, et al. Expression and regulation of transcription factors GATA-4 and GATA-6 in developing mouse testis[J]. Endocrinology, 1999, 140(3): 1470–1480. DOI: 10.1210/endo.140.3.6587 |
| [24] | SILVERMAN E, EIMERL S, ORLY J. CCAAT enhancer-binding protein beta and GATA-4 binding regions within the promoter of the steroidogenic acute regulatory protein (StAR) gene are required for transcription in rat ovarian cells[J]. J Biol Chem, 1999, 274(25): 17987–17996. DOI: 10.1074/jbc.274.25.17987 |
| [25] | TREMBLAY J J, HAMEL F, VIGER R S. Protein kinase A-dependent cooperation between GATA and CCAAT/enhancer-binding protein transcription factors regulates steroidogenic acute regulatory protein promoter activity[J]. Endocrinology, 2002, 143(10): 3935–3945. DOI: 10.1210/en.2002-220413 |
| [26] | LAVOIE H A, SINGH D, HUI Y Y. Concerted regulation of the porcine steroidogenic acute regulatory protein gene promoter activity by follicle-stimulating hormone and insulin-like growth factor Ⅰ in granulosa cells involves GATA-4 and CCAAT/Enhancer binding protein β[J]. Endocrinology, 2004, 145(7): 3122–3134. DOI: 10.1210/en.2003-1719 |
| [27] | ROBERT N M, TREMBLAY J J, VIGER R S. Friend of GATA (FOG)-1 and FOG-2 differentially repress the GATA-dependent activity of multiple gonadal promoters[J]. Endocrinology, 2002, 143(10): 3963–3973. DOI: 10.1210/en.2002-220280 |
| [28] |
苟华. GATA-6过表达和干扰载体的构建及其对鹅颗粒细胞类固醇合成及相关基因的调控研究[D]. 雅安: 四川农业大学, 2015.
GOU H. Construction of GATA-6 eukaryotic expression and shRNA interference vectors to characterize its role on steroidogenesis in goose granulosa cells[D]. Ya'an: Sichuan Agricultural University, 2015. (in Chinese) |
| [29] | YAN Y E, LIU L, WANG J F, et al. Prenatal nicotinic exposure suppresses fetal adrenal steroidogenesis via steroidogenic factor 1(SF-1) deacetylation[J]. Toxicol Appl Pharmacol, 2014, 277(3): 231–241. DOI: 10.1016/j.taap.2014.03.019 |
| [30] | LAKSHMANAN M, MOHANTY B, LIM S H, et al. Metabolic and transcriptional regulatory mechanisms underlying the anoxic adaptation of rice coleoptile[J]. AoB Plants, 2014, 6: plu026. |
| [31] | MOHANTY B, LAKSHMANAN M, LIM S H, et al. Light-specific transcriptional regulation of the accumulation of carotenoids and phenolic compounds in rice leaves[J]. Plant Signal Behav, 2016, 11(6): e1184808. DOI: 10.1080/15592324.2016.1184808 |

 图 1(Figure 1)
图 1(Figure 1)


