中国地方绵羊品种根据尾型分为5类:长脂尾、短脂尾、短瘦尾、长瘦尾和脂臀尾[1]。将脂肪沉积在尾部这是绵羊独有的特性。尾部脂肪在逆境条件下(如寒冷等)提供身体能量所需,类似于骆驼的驼峰,属于适应性性状[2]。但是随着肥胖、心脑血管以及糖尿病的增加,人们更倾向于高蛋白和低脂肪的肉类[3]。另外,从饲养成本来看,生产1 kg脂肪所消耗的饲料可产生2 kg瘦肉[4],因此,尾脂沉积过多不利于产生经济效益。
不同尾型的绵羊品种是通过长期的人工选择和自然选择形成的,而选择也会在基因组上留下烙印,这就是选择信号(selection signature,SS)[5]。目前,选择信号检测方法很多[6],其中对两个及多个群体进行选择信号检测时比较经典的方法就是FST[7],该方法也常应用于羊的选择信号检测中[3, 8-13]。在现有文献资料中,对肥瘦尾性状之间的选择信号检测时[3, 11-13],都是将不同品种的羊简单的按照尾型划分为肥、瘦尾两类,这样就很难消除不同遗传背景对检测结果的影响。其实在中国地方品种中,呼伦贝尔羊(属于短脂尾绵羊品种)群体内存在明显的尾部大小差异,主要是因为该品种内存在两个品系[14],这为同一个品种内不同品系开展选择信号检测提供了一个理想的动物资源。
本研究使用Illumina Ovine 600K BeadChip高密度SNP芯片,针对呼伦贝尔羊不同品系利用可变窗口FST进行选择信号检测。旨在挖掘呼伦贝尔羊尾部脂肪沉积的候选基因,揭示影响绵羊尾部脂肪沉积基因,为培育短尾绵羊品种提供理论依据。
1 材料与方法 1.1 试验材料在内蒙古呼伦贝尔草原的牧场中,从6月龄的呼伦贝尔羊群随机挑选2 000只绵羊,然后选出大尾和小尾各150只,并对活体绵羊进行尾部长度、尾部最宽处的长度和周长及体尺数据进行测量,对所选个体查找系谱信息,使用一次性含有EDTA抗凝的真空采血管进行颈静脉采血,-20 ℃冷冻保存,用于基因组DNA提取。
1.2 试验方法 1.2.1 基因型检测首先利用血液基因组DNA提取试剂盒(天根生化科技有限公司)提取血液样品中的基因组DNA;定制Illumina Ovine SNP600K芯片,获得601 715个SNP分型数据;通过GenomeStudio软件进行数据删选以及SNP等位基因型频率的分析,利用PLINK软件[15]进行质量控制。质控的标准是:平均检出率大于0.95;平均最小等位基因频率大于0.02;剔除未定位的和性染色体上的SNPs位点。通过质控后,剩余577 401个SNPs,大尾羊142只,小尾羊146只。再利用Beagle软件[16]对缺失数据进行填充,用于后续的分析。
1.2.2 选择信号检测方法本研究采用群体分化系数FST法进行呼伦贝尔羊品种内不同品系间选择信号检测。群体分化指数FST可用来度量群体分化程度,进而估计品系间的分化程度。本研究利用PLINK软件[15]计算每个SNP位点的FST值,其计算基本原理是按照Weir和Cockerham[17]描述的无偏估计方法,其计算公式:
| $ {F_{ST}} = \frac{{MSP - MSG}}{{MSP + ({n_C} - 1)MSG}} $ | (1) |
其中,MSG为检测的群体内部位点的误差均方,其计算公式:
| $ MSG = \frac{1}{{\sum\nolimits_{i = 1}^1 {({n_i} - 1)} }}\sum\nolimits_{i = 1}^s {{n_i}{P_{{A_i}}}(1 - {P_{{A_i}}})} $ | (2) |
MSP是检测的群体之间位点的均方差,其计算公式:
| $ MSP = \frac{1}{{S - 1}}\sum\nolimits_{i = 1}^S {{n_i}{{({P_{{A_i}}} - {{\bar P}_A})}^2}} $ | (3) |
nC指校正后群体间的平均样本大小,其计算公式:
| $ {n_C} = \frac{1}{{S - 1}}\left[ {\sum\nolimits_{i = 1}^S {{n_i}} - \frac{{\sum\nolimits_{i = 1}^S {n_i^2} }}{{\sum\nolimits_{i = 1}^S {{n_i}} }}} \right] $ | (4) |
上述各式中,i是总亚群数S的一个群体,i=1, 2, …, S;PAi是第i个亚群中SNP等位基因A的频率;ni是亚群体i的平均样本大小;PA是各群体中PA的加权平均值,即
通过公式(1)计算出每个位点的FST值。再根据每条染色体上每个SNP的FST值大小利用三次光滑样条估计方法确定窗口大小和数量[18],主要利用R语言中GenWin包完成。然后通过整个基因组范围内的每个窗口平均FST值和SNP个数,构建全基因组范围的W统计量:
| $ W = \frac{{\left( {\bar X - \mu } \right)}}{{\sqrt {{s^2}/n} }} $ | (5) |
其中,X表示一个窗口内FST的平均值,μ表示的是整个基因组范围内FST的平均值,s2表示全基因组范围内FST的样本方差,n表示一个窗口内的SNPs数目。
1.2.3 候选基因的检测和注释参照NCBI数据库(https://www.ncbi.nlm.nih.gov/)和CSIRO数据库(https://www.livestockgenomics.csiro.au/sheep/oar3.1.php)的Ovis aries 3.1基因组信息,对筛选出来的受选择位点进行基因注释。以选择信号发生区域核心SNP为中心,上下游各扩展1 000 kb为选择区段。将落在这个选择区段内的基因定义为选择信号的“候选基因”。
1.2.4 生物信息学分析利用Gorilla[19](http://cbl-gorilla.cs.technion.ac.il/)进行分析,包括细胞组分(cellular component)、分子功能(molecular function)和生物学过程(biological process)分析。
本研究采用R语言[20](https://www.r-project.org/)进行数据处理、分析以及试验结果的整理。
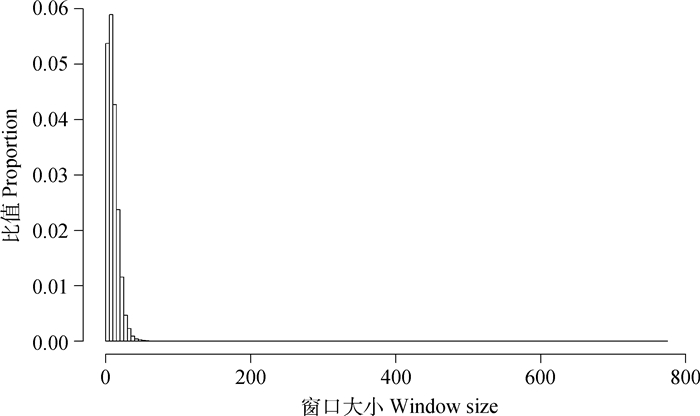
2 结果 2.1 呼伦贝尔羊全基因组范围内可变窗口分布通过三次光滑样条估计方法确定基因组范围内23 144个可变窗口。窗口包含的SNPs个数从1到775个,其中包含最多数目的SNP窗口位于chr12: 35 241 750~43 798 950 bp处,该窗口大小为8.56 Mb。图 1展示了窗口内SNP频数直方,其中包括≤30个SNPs的窗口数所占总窗口数的97.7%。
|
图 1 窗口内SNP频数直方图 Figure 1 Histogram of the SNPs frequency within windows |
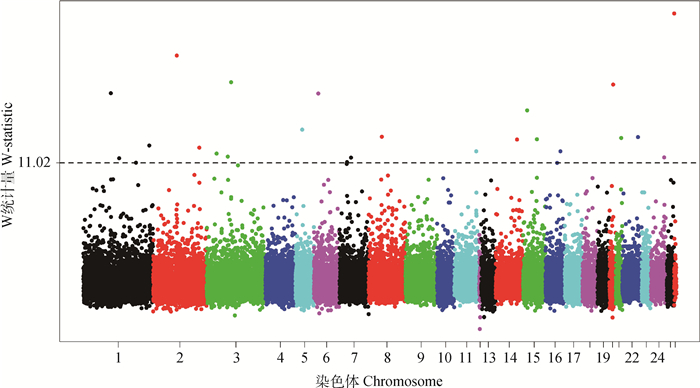
呼伦贝尔羊全基因组范围内的W统计量分布情况见图 2,以W统计量为11.02作为阈值,即P < 0.001,共检测出27个显著的基因区段。按照W统计量的大小列在表 1中,主要分布在1、2、3、6、7、11、14、15、20、21、22和24号染色体上,最显著的区段位于26号染色体上。
|
图 2 呼伦贝尔羊全基因组W统计量值分布 Figure 2 Genome-wide distribution of W-statistic values in Hulun Buir sheep |
|
|
表 1 W统计量筛选的选择区段及候选基因 Table 1 Selected regions identified by W-statistic and annotated candidate genes |
通过基因注释,在27个显著区段内共找到了337个候选基因(表 1)。其中最显著的区段Chr26: 0.00~0.11,找到了两个候选基因:ERICHI、DLGAP2。ERICHI基因功能目前报道的比较少,而DLGAP2报道与神经元突触功能有关,是精神分裂症的易感基因[21]。通过数据库查找发现与脂代谢相关的基因只有22个(表 2)。
|
|
表 2 筛选出已报道与脂肪合成代谢相关的候选基因 Table 2 The candidate genes related to lipid metabolism in previous studies |
本研究通过在线工具Gorilla(http://cbl-gorilla.cs.technion.ac.il/),将FDR阈值设为0.001,对获得的337个候选基因进行基因的GO富集分析。分析结果见表 3,从表 3可以看出受到选择的基因主要集中在小分子代谢过程、有机氮化合物代谢过程等生物学过程。在细胞组分上主要集中在细胞内组成成分上,总共发现了197个基因,占总基因的58.46%,在所有富集条目中数量占比最多。而分子功能富集分析没有发现受选择基因。
|
|
表 3 选择区段内基因GO富集条目汇总 Table 3 Summary of enrichment analysis of GO terms for genes in selected regions |
因为相同染色体上的SNPs之间受到连锁不平衡和重组率等因素的影响,在选择信号检测时,经常将基因组分成等距离区间,然后计算每个区间内所有单点FST的平均值,将其作为整个区间的值。一般区间划分方法有两种:一种就是固定窗口,另外一种就是滑动窗口。固定窗口就是将整个基因组分成相同片段的区间,而滑动窗口就是相邻窗口之间有一定的重合。但是,第一种方法对窗口大小确定具有一定的随机性和主观性,而第二种方法使得相邻区间统计值之间又具有很强的相关性,因此,这两种方法都会降低检测功效。于是Beissinger等[18]提出三次光滑样条估计方法,为确定窗口大小和数量的新方法。该方法不仅可以消除噪音的干扰,又不会减低检测信号的功效。这也是本研究采用可变窗口FST值的主要原因。
3.2 呼伦贝尔羊品系间差异分析呼伦贝尔羊由巴尔虎品系(半椭圆状尾)和短尾品系(小桃状尾)组成,主要分布在内蒙古的呼伦贝尔牧区草原上[14]。短尾品系的形成可能与引入了前苏联的后贝加尔羊血液有关[22]。目前两个品系除了在尾型上存在很大差异外,其他外貌特征基本上一致[14]。为了能更好的区分这两个品系,本研究对尾部脂肪重量测量后选择两端重量的个体进行分析,使得检测结果更加准确。
3.3 与前期研究结果的比较与前期研究[3, 11-13]相比,本研究采用了不同的群体、试验设计、检测方法和极显著的判断标准(P < 0.001),所以并没有找到与前期研究完全重合的基因区段,但也找到了与前期研究临近的显著区段。Chr3: 94.05~94.21 Mb与Yuan等[11]研究所检测的区段Chr3: 93.33~93.98 Mb相距0.07 Mb;Chr22: 40.92~41.01 Mb与刘真等[3]在显著区段Chr22: 40.23~40.73 Mb相距0.19 Mb。此外,在Chr1: 140.59~140.69 Mb区段也验证了Xu等[23]通过GWAS找到的与绵羊尾部脂肪沉积相关的候选基因NRIP1(又叫RIP140)。因此,这些区段所对应的基因或者鉴定的基因可作为控制绵羊尾脂重要的候选基因。
3.4 脂肪沉积相关基因功能讨论本研究检测到了337个候选基因,有22个基因在已报道文献中与脂肪代谢相关。其中与猪脂肪代谢相关的基因有IGFBP2、AKT2、LIPE、MDH2和NPNT。IGFBP2可在脂肪组织中表达,与猪眼肌面积、肥肉率、瘦肥比、花油和色值等胴体和肉质性状显著相关[24-26]。AKT2可以调节肌肉与脂肪细胞内的葡萄糖转运[27],也与猪脂肪沉积相关[28]。LIPE是一种脂肪酶,可影响金华猪胴体脂肪的合成[29]。MDH2在三羧酸循环中可催化苹果酸生成丙酮酸,并产生还原型辅酶Ⅱ(NADPH),而NADPH是动物体内脂肪酸合成及碳链延长过程中的重要辅酶和脂肪酸合成的供氢体,体内NADPH的供应情况直接影响脂肪及类脂的合成。另外,Zhou等[30]研究发现,MDH2基因在荣昌猪不同部位脂肪组织中存在差异化表达。NPNT基因可以有效降低肉牛肌内脂肪含量和大理石花纹的密度[31]。由PRLR介导的PRL可直接调节脂肪组织中的代谢过程[32-33]。解偶联蛋白-2(UCP2)是线粒体转运蛋白超家族的成员,在脂肪组织中高度表达,而脂肪酸进入脂肪组织的总量又可调节UCP2基因的表达[34-36]。另外,褐色脂肪组织UCP1具有明显的解偶联活性,对于维持小型哺乳动物的体温至关重要[37]。NRIP1基因可作为调控脂粒和葡萄糖代谢的重要因子[38-40]。ZNF638是脂肪生成的新型和早期调节剂[41]。TMEFF2基因在白色脂肪组织中高度表达[42]。ACOX1是脂肪细胞内参与脂肪酸氧化相关的酶[43]。DDB1直接与SIRT7结合,从而抑制E3泛素连接酶复合物的形成,最终导致参与脂肪代谢的核受体TR4含量增加[44]。DDB1还参与组成WDTC1的一个结合元件,而WDTC1可调节脂肪相关基因的转录,其功能的丧失会导致脂肪细胞增加,从而引起脂肪沉积[45]。RYR3基因是影响中国人群循环脂联素的QTL,而脂联素是脂肪细胞分泌的细胞因子,可影响脂代谢和糖代谢[46]。
与奶牛乳脂相关的基因有FADS1、FADS2和ACADSB。脂肪酸脱氢酶1(FADS1)和脂肪酸脱氢酶2(FADS2)是多不饱和脂肪酸合成途径的关键酶。FADS1基因与奶牛乳脂率[47]和鸡肉脂肪含量[48]密切相关。FADS2中的rs174575基因多态性与乳汁ALA水平相关联[49]。而且FADS1/2基因的多态性与脂肪酸代谢和脂肪组织炎症相关,最终会导致体重减轻[50-51]。短支链酰基辅酶A脱氢酶(ACADSB)在乳脂代谢中起到重要作用,对细胞中脂肪含量和甘油三酯代谢起到调控作用[52]。
另外还有一些基因与脂肪代谢相关的疾病有关。CD1D、ABCG1、SAP30BP、RAB37和ARAP1与2型糖尿病相关。2型糖尿病患者因为胰岛素功能不足,使糖代谢功能减弱,脂肪沉积增加,最终导致肥胖[53]。脂肪细胞CD1D在脂肪iNKT细胞的刺激中起关键作用,可导致高脂饮食小鼠抗炎反应[54],也可破坏肥胖小鼠内脏脂肪组织中的免疫平衡,最终引起2型糖尿病的发生[55]。三磷酸结合盒转运体G1(ATP-binding cassette G1,ABCG1)在胆固醇逆向转运过程中发挥着重要作用,并参与脂肪代谢[56],ABCG1与动脉粥样硬化[57]、肥胖[58-59]、糖尿病[60]等代谢疾病密切相关。Chen等[61]通过单个组织中表达数量性状位点(eQTL)鉴定出SAP30BP是肥胖和2型糖尿病的候选基因。RAB37基因甲基化之后容易造成成糖化血红蛋白(HbA1c)生成,而糖化血红蛋白与2型糖尿病、肥胖等疾病有关,RAB37基因间接影响着脂肪组织的代谢[62]。ARAP1表达量增加会导致胰岛素功能降低,从而导致患2型糖尿病的风险增加[63]。
4 结论本研究利用高密度SNP绵羊芯片,针对呼伦贝尔羊品种内不同品系开展全基因组选择信号检测,并利用最新提出的三次光滑样条估计方法确定窗口大小和数量,共检测出27个受选择区段,其中3个区段验证了已有的报道。这些区段注释了337个候选基因,其中有22个候选基因在相关文献中已报道与脂肪代谢相关。这些候选基因主要富集在细胞内组成成分、有机氮化合物代谢过程以及小分子代谢过程等条目中。这为绵羊尾型选育、品种改良以及后续功能基因验证提供了重要依据。
| [1] |
国家畜禽遗传资源委员会.
中国畜禽遗传资源志·羊志[M]. 北京: 中国农业出版社, 2011.
China National Commission of Animal Genetic Resources. Animal genetic resources in China? Sheep and goat[M]. Beijing: Chinese Agricultural Press, 2011. (in Chinese) |
| [2] |
甘尚权, 张伟, 宋天增, 等. X染色体一处新发现的SNP位点在脂尾(臀)、瘦尾绵羊群体中的多态检测及分析[J]. 西南农业学报, 2013, 26(5): 2066–2070.
GAN S Q, ZHANG W, SONG T Z, et al. Polymorphism detection and analysis of novel SNP on X chromosome between fat-tailed and thin-tailed sheep breeds[J]. Southwest China Journal of Agricultural Sciences, 2013, 26(5): 2066–2070. (in Chinese) |
| [3] |
刘真, 王慧华, 刘瑞凿, 等. 不同尾型绵羊全基因组选择信号检测[J]. 畜牧兽医学报, 2015, 46(10): 1721–1732.
LIU Z, WANG H H, LIU R Z, et al. Genome-wide detection of selection signatures of distinct tail types in sheep populations[J]. Acta Veterinaria et Zootechnica Sinica, 2015, 46(10): 1721–1732. (in Chinese) |
| [4] |
甘尚权, 张伟, 沈敏, 等. 绵羊X染色体59383635位点多态性与脂尾性状的相关性分析[J]. 遗传, 2013, 35(10): 1209–1216.
GAN S Q, ZHANG W, SHEN M, et al. Correlation analysis between polymorphism of the 59383635th locus on X chromosome and fat-tail trait in sheep[J]. Hereditas, 2013, 35(10): 1209–1216. (in Chinese) |
| [5] |
马云龙, 张勤, 丁向东. 利用高密度SNP检测不同猪品种间X染色体选择信号[J]. 遗传, 2012, 34(10): 1251–1260.
MA Y L, ZHANG Q, DING X D. Detecting selection signatures on X chromosome in pig through high density SNPs[J]. Hereditas, 2012, 34(10): 1251–1260. (in Chinese) |
| [6] | NIELSEN R. Molecular signatures of natural selection[J]. Annu Rev Genet, 2005, 39: 197–218. DOI: 10.1146/annurev.genet.39.073003.112420 |
| [7] | HOLSINGER K E, WEIR B S. Genetics in geographically structured populations:defining, estimating and interpreting FST[J]. Nat Rev Genet, 2009, 10(9): 639–650. DOI: 10.1038/nrg2611 |
| [8] |
曾滔, 赵福平, 王光凯, 等. 基于群体分化指数FST的绵羊全基因组选择信号检测[J]. 畜牧兽医学报, 2013, 44(12): 1891–1899.
ZENG T, ZHAO F P, WANG G K, et al. Genome-wide detection of selection signatures in sheep populations with use of population differentiation index FST[J]. Acta Veterinaria et Zootechnica Sinica, 2013, 44(12): 1891–1899. (in Chinese) |
| [9] | KIJAS J W, LENSTRA J A, HAYES B, et al. Genome-wide analysis of the world's sheep breeds reveals high levels of historic mixture and strong recent selection[J]. PLoS Biol, 2012, 10(2): e1001258. DOI: 10.1371/journal.pbio.1001258 |
| [10] | GORKHALI N A, DONG K Z, YANG M, et al. Genomic analysis identified a potential novel molecular mechanism for high-altitude adaptation in sheep at the Himalayas[J]. Sci Rep, 2016, 6: 29963. DOI: 10.1038/srep29963 |
| [11] | YUAN Z H, LIU E, LIU Z, et al. Selection signature analysis reveals genes associated with tail type in Chinese indigenous sheep[J]. Anim Genet, 2017, 48(1): 55–66. DOI: 10.1111/age.2017.48.issue-1 |
| [12] | MOIOLI B, PILLA F, CIANI E. Signatures of selection identify loci associated with fat tail in sheep[J]. J Anim Sci, 2015, 93(10): 4660–4669. DOI: 10.2527/jas.2015-9389 |
| [13] | MORADI M H, NEJATI-JAVAREMI A, MORADI-SHAHRBABAK M, et al. Genomic scan of selective sweeps in thin and fat tail sheep breeds for identifying of candidate regions associated with fat deposition[J]. BMC Genet, 2012, 13: 10. |
| [14] |
任鑫亮, 高雅英. 呼伦贝尔羊的特性及饲养管理措施[J]. 畜牧与饲料科学, 2012, 33(3): 122–123.
REN X L, GAO Y Y. Characteristics of Hulun Buir Sheep and breeding management measures[J]. Animal Husbandry and Feed Science, 2012, 33(3): 122–123. (in Chinese) |
| [15] | PURCELL S, NEALE B, TODD-BROWN K, et al. PLINK:a tool set for whole-genome association and population-based linkage analyses[J]. Am J Hum Genet, 2007, 81(3): 559–575. DOI: 10.1086/519795 |
| [16] | BROWNING B L, BROWNING S R. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals[J]. Am J Hum Genet, 2009, 84(2): 210–223. DOI: 10.1016/j.ajhg.2009.01.005 |
| [17] | WEIR B S, COCKERHAM C C. Estimating F-statistics for the analysis of population structure[J]. Evolution, 1984, 38(6): 1358–1370. |
| [18] | BEISSINGER T M, ROSA G J M, KAEPPLER S M, et al. Defining window-boundaries for genomic analyses using smoothing spline techniques[J]. Genet Sel Evol, 2015, 47(1): 30. DOI: 10.1186/s12711-015-0105-9 |
| [19] | EDEN E, NAVON R, STEINFELD I, et al. GOrilla:a tool for discovery and visualization of enriched GO terms in ranked gene lists[J]. BMC Bioinformatics, 2009, 10: 48. DOI: 10.1186/1471-2105-10-48 |
| [20] | R CORE TEAM. R: A language and environment for statistical computing[M]. Vienna, Austria: the R Foundation, 2015, 14: 12-21. |
| [21] | LI J M, LU C L, CHENG M C, et al. Role of the DLGAP2 gene encoding the SAP90/PSD-95-associated protein 2 in schizophrenia[J]. PLoS One, 2014, 9(1): e85373. DOI: 10.1371/journal.pone.0085373 |
| [22] |
李宏图, 秦秀娟, 乌恩巴雅尔, 等. 呼伦贝尔羊"短尾"品系的外形分析[J]. 中国草食动物, 2006(S1): 148–149.
LI H T, QIN X J, WUEN B Y E, et al. Analysis of shape of short-tailed Hulun Buir sheep[J]. China Herbivores, 2006(S1): 148–149. (in Chinese) |
| [23] | XU S S, REN X, YANG G L, et al. Genome-wide association analysis identifies the genetic basis of fat deposition in the tails of sheep (Ovis aries)[J]. Anim Genet, 2017, 48(5): 560–569. DOI: 10.1111/age.2017.48.issue-5 |
| [24] | SABIN M A, YAU S W, RUSSO V C, et al. Dietary monounsaturated fat in early life regulates IGFBP2:implications for fat mass accretion and insulin sensitivity[J]. Obesity, 2011, 19(12): 2374–2381. DOI: 10.1038/oby.2011.55 |
| [25] |
杨华, 徐珍, 左波. 猪IGFBP2基因多态性及其与胴体、肉质性状的关联分析[J]. 中国畜牧杂志, 2017, 53(3): 25–28.
YANG H, XU Z, ZUO B. Polymorphism of IGFBP2 gene and its association with carcass and meat quality traits in pigs[J]. Chinese Journal of Animal Science, 2017, 53(3): 25–28. (in Chinese) |
| [26] | GENG T Y, SUTTER A, HARLAND M D, et al. SphK1 mediates hepatic inflammation in a mouse model of NASH induced by high saturated fat feeding and initiates proinflammatory signaling in hepatocytes[J]. J Lipid Res, 2015, 56(12): 2359–2371. DOI: 10.1194/jlr.M063511 |
| [27] | LIU Z J, GAN L, LIU G N, et al. Sirt1 decreased adipose inflammation by interacting with Akt2 and inhibiting mTOR/S6K1 pathway in mice[J]. J Lipid Res, 2016, 57(8): 1373–1381. DOI: 10.1194/jlr.M063537 |
| [28] | OTIENO C J, BASTIAANSEN J, RAMOS A M, et al. Mapping and association studies of diabetes related genes in the pig[J]. Anim Genet, 2005, 36(1): 36–42. DOI: 10.1111/age.2005.36.issue-1 |
| [29] | MIAO Z, ZHU F, ZHANG H, et al. Developmental patterns of FASN and LIPE mRNA expression in adipose tissue of growing Jinhua and Landrace gilts[J]. Czech J Anim Sci, 2010, 55(12): 557–564. DOI: 10.17221/CJAS |
| [30] | ZHOU S L, LI M Z, LI Q H, et al. Differential expression analysis of porcine MDH1, MDH2 and ME1 genes in adipose tissues[J]. Genet Mol Res, 2012, 11(2): 1254–1259. DOI: 10.4238/2012.May.9.4 |
| [31] | CLARK D L. Transcription profiles of differentially marbled beef cattle[D]. Champaign: University of Illinois at Urbana-Champaign, 2008. |
| [32] | LING C, HELLGREN G, GEBRE-MEDHIN M, et al. Prolactin (PRL) receptor gene expression in mouse adipose tissue:increases during lactation and in PRL-transgenic mice[J]. Endocrinology, 2000, 141(10): 3564–3572. DOI: 10.1210/endo.141.10.7691 |
| [33] | LING C, SVENSSON L, ODÉN B, et al. Identification of functional prolactin (PRL) receptor gene expression:PRL inhibits lipoprotein lipase activity in human white adipose tissue[J]. J Clin Endocrinol Metab, 2003, 88(4): 1804–1808. DOI: 10.1210/jc.2002-021137 |
| [34] | AUBERT J, CHAMPIGNY O, SAINT-MARC P, et al. Up-regulation of UCP-2 gene expression by PPAR agonists in preadipose and adipose cells[J]. Biochem Biophys Res Commun, 1997, 238(2): 606–611. DOI: 10.1006/bbrc.1997.7348 |
| [35] | ESTERBAUER H, SCHNEITLER C, OBERKOFLER H, et al. A common polymorphism in the promoter of UCP2 is associated with decreased risk of obesity in middle-aged humans[J]. Nat Genet, 2001, 28(2): 178–183. DOI: 10.1038/88911 |
| [36] | PEDERSEN S B, BRUUN J M, KRISTENSEN K, et al. Regulation of UCP1, UCP2, and UCP3 mRNA expression in brown adipose tissue, white adipose tissue, and skeletal muscle in rats by estrogen[J]. Biochem Biophys Res Commun, 2001, 288(1): 191–197. DOI: 10.1006/bbrc.2001.5763 |
| [37] | RICQUIER D, BOUILLAUD F. The uncoupling protein homologues:UCP1, UCP2, UCP3, StUCP and AtUCP[J]. Biochem J, 2000, 345(2): 161–179. |
| [38] | HO P C, CHUANG Y S, HUNG C H, et al. Cytoplasmic receptor-interacting protein 140(RIP140) interacts with perilipin to regulate lipolysis[J]. Cell Signalling, 2011, 23(8): 1396–1403. DOI: 10.1016/j.cellsig.2011.03.023 |
| [39] | HOCHBERG I, TRAN Q T, BARKAN A L, et al. Gene expression signature in adipose tissue of acromegaly patients[J]. PLoS One, 2015, 10(6): e0129359. DOI: 10.1371/journal.pone.0129359 |
| [40] | LEONARDSSON G, STEEL J H, CHRISTIAN M, et al. Nuclear receptor corepressor RIP140 regulates fat accumulation[J]. Proc Natl Acad Sci U S A, 2004, 101(22): 8437–8442. DOI: 10.1073/pnas.0401013101 |
| [41] | MERUVU S, HUGENDUBLER L, MUELLER E. Regulation of adipocyte differentiation by the zinc finger protein ZNF638[J]. J Biol Chem, 2011, 286(30): 26516–26523. DOI: 10.1074/jbc.M110.212506 |
| [42] | CHEN T R, WANG P, CARROLL L K, et al. Generation and characterization of Tmeff2 mutant mice[J]. Biochem Biophys Res Commun, 2012, 425(2): 189–194. DOI: 10.1016/j.bbrc.2012.07.064 |
| [43] | OSUMI T, HASHIMOTO T. Acyl-CoA oxidase of rat liver:a new enzyme for fatty acid oxidation[J]. Biochem Biophys Res Commun, 1978, 83(2): 479–485. DOI: 10.1016/0006-291X(78)91015-X |
| [44] | KARIM M F, YOSHIZAWA T, SOBUZ S U, et al. Sirtuin 7-dependent deacetylation of DDB1 regulates the expression of nuclear receptor TR4[J]. Biochem Biophys Res Commun, 2017, 490(2): 423–428. DOI: 10.1016/j.bbrc.2017.06.057 |
| [45] | DUCOS E, VERGōS V, DE BERNONVILLE T D, et al. Remarkable evolutionary conservation of antiobesity ADIPOSE/WDTC1 homologs in animals and plants[J]. Genetics, 2017, 207(1): 153–162. DOI: 10.1534/genetics.116.198382 |
| [46] | CHANG Y C, CHIU Y F, HE C T, et al. Genome-wide linkage analysis and regional fine mapping identified variants in the RYR3 gene as a novel quantitative trait locus for circulating adiponectin in Chinese population[J]. Medicine, 2016, 95(44): e5174. DOI: 10.1097/MD.0000000000005174 |
| [47] |
徐晨希, 王梦琦, 朱小瑞, 等. 中国荷斯坦牛FADS2基因3'端SNP突变对乳中脂肪酸组成的影响[J]. 中国农业科学, 2016, 49(11): 2194–2202.
XU C X, WANG M Q, ZHU X R, et al. Effects of SNPs in the 3' untranslated regions of FADS2 on the composition of fatty acids in milk of Chinese Holstein[J]. Scientia Agricultura Sinica, 2016, 49(11): 2194–2202. DOI: 10.3864/j.issn.0578-1752.2016.11.015 (in Chinese) |
| [48] | BOSCHETTI E, BORDONI A, MELUZZI A, et al. Fatty acid composition of chicken breast meat is dependent on genotype-related variation of FADS1 and FADS2 gene expression and desaturating activity[J]. Animal, 2016, 10(4): 700–708. DOI: 10.1017/S1751731115002712 |
| [49] |
丁珍, 李响, 吴义霞, 等. FADS2基因rs174575多态性与乳汁多不饱和脂肪酸水平的关系研究[J]. 中国妇幼保健, 2015, 30(20): 3464–3466.
DING Z, LI X, WU Y X, et al. Study on the relationship between rs174575 polymorphism of FADS2 gene and polyunsaturated fatty acids levels in breast milk[J]. Maternal & Child Health Care of China, 2015, 30(20): 3464–3466. (in Chinese) |
| [50] | VAITTINEN M, MÄNNISTÖ V, KÄKELÄ P, et al. Interorgan cross talk between fatty acid metabolism, tissue inflammation, and FADS2 genotype in humans with obesity[J]. Obesity, 2017, 25(3): 545–552. DOI: 10.1002/oby.21753 |
| [51] | VAITTINEN M, WALLE P, KUOSMANEN E, et al. FADS2 genotype regulates delta-6 desaturase activity and inflammation in human adipose tissue[J]. J Lipid Res, 2016, 57(1): 56–65. DOI: 10.1194/jlr.M059113 |
| [52] | ENSENAUER R, HE M, WILLARD J M, et al. Human acyl-CoA dehydrogenase-9 plays a novel role in the mitochondrial β-oxidation of unsaturated fatty acids[J]. J Biol Chem, 2005, 280(37): 32309–32316. DOI: 10.1074/jbc.M504460200 |
| [53] | KUWABARA W M T, PANVELOSKI-COSTA A C, YOKOTA C N F, et al. Comparison of Goto-Kakizaki rats and high fat diet-induced obese rats:Are they reliable models to study Type 2 Diabetes mellitus?[J]. PLoS One, 2017, 12(12): e0189622. DOI: 10.1371/journal.pone.0189622 |
| [54] | HUH J Y, PARK J, KIM J I, et al. Deletion of CD1d in adipocytes aggravates adipose tissue inflammation and insulin resistance in obesity[J]. Diabetes, 2017, 66(4): 835–847. DOI: 10.2337/db16-1122 |
| [55] | ZHANG H, XUE R, ZHU S, et al. M2-specific reduction of CD1d switches NKT cell-mediated immune responses and triggers metaflammation in adipose tissue[J]. Cell Mol Immunol, 2017. DOI: 10.1038/cmi.2017.11 |
| [56] | MA Z S, SONG Z Y, ZHANG Q. Cholesterol efflux is LXRα isoform-dependent in human macrophages[J]. BMC Cardiovasc Disord, 2014, 14: 80. DOI: 10.1186/1471-2261-14-80 |
| [57] | LANG J K, CIMATO T R. Cholesterol and hematopoietic stem cells:inflammatory mediators of atherosclerosis[J]. Stem Cell Transl Med, 2014, 3(5): 549–552. DOI: 10.5966/sctm.2013-0205 |
| [58] | BATES S R, TAO J Q, COLLINS H L, et al. Pulmonary abnormalities due to ABCA1 deficiency in mice[J]. Am J Physiol Lung Cell Mol Physiol, 2005, 289(6): L980–L989. DOI: 10.1152/ajplung.00234.2005 |
| [59] | EDGEL K A, MCMILLEN T S, WEI H, et al. Obesity and weight loss result in increased adipose tissue ABCG1 expression in db/db mice[J]. Biochim Biophys Acta, 2012, 1821(3): 425–434. DOI: 10.1016/j.bbalip.2011.11.012 |
| [60] | SPARTANO N L, LAMON-FAVA S, MATTHAN N R, et al. Regulation of ATP-binding cassette transporters and cholesterol efflux by glucose in primary human monocytes and murine bone marrow-derived macrophages[J]. Exp Clin Endocrinol Diabetes, 2014, 122(8): 463–468. DOI: 10.1055/s-00000017 |
| [61] | CHEN J, MENG Y, ZHOU J, et al. Identifying candidate genes for Type 2 Diabetes Mellitus and obesity through gene expression profiling in multiple tissues or cells[J]. J Diabetes Res, 2013, 2013: 970435. |
| [62] | RÖNN T, VOLKOV P, GILLBERG L, et al. Impact of age, BMI and HbA1c levels on the genome-wide DNA methylation and mRNA expression patterns in human adipose tissue and identification of epigenetic biomarkers in blood[J]. Hum Mol Genet, 2015, 24(13): 3792–3813. |
| [63] | KULZER J R, STITZEL M L, MORKEN M A, et al. A common functional regulatory variant at a Type 2 diabetes locus upregulates ARAP1 expression in the pancreatic beta cell[J]. Am J Hum Genet, 2014, 94(2): 186–197. DOI: 10.1016/j.ajhg.2013.12.011 |



