猪肉是人民膳食结构的主要组成部分,据美国农业部经济学家和经合组织-粮农组织预测,与2014年相比,到2023年全球猪肉产量将会至少增加10.5%[1]。随着人们需求量的增加,对猪肉肉质的要求也越来越高,主要包括肉色、嫩度、风味和多汁性等。肌内脂肪是影响肉质嫩度、风味以及多汁性的重要因素[2-3],有研究发现,某些生物活性物质如共轭亚油酸在提高肌内脂肪含量的同时,却降低或维持了皮下脂肪的沉积量[4],由此推测,肌内脂肪可能具有不同于皮下脂肪的生物学功能以及分子调控机制。因此,了解猪肉肌内脂肪沉积的分子机制有助于改善猪肉品质。
环状RNA(circular RNA, circRNA)是一类由特殊的选择性剪切产生的环形内源性分子,呈共价闭合的环形结构,没有5′和3′极性,不能够被核糖核酸酶降解。在哺乳动物细胞中,circRNA分布广泛,含量丰富,且具有稳定性、保守性和组织特异性。已有研究发现,circRNA与阿尔茨海默病[5]、大肠癌和卵巢癌[6]、肝细胞癌[7]、心血管疾病[8]、中枢神经紊乱[9]等疾病相关,除此之外,还与细胞增殖[10]、卵子发生与受精卵的形成[11]有关。在真核细胞中,circRNA可以作为miRNA海绵来调控基因表达[12-13]。研究发现,miRNA和多种转录因子参与调控与脂肪细胞分化和脂质代谢相关基因的表达[14-18]。但是,关于circRNA是否参与调控脂肪沉积和脂代谢还未见报道。
莱芜猪肉质优良,其肌内脂肪含量高达10.32%[19];而大白猪属于典型的瘦肉型猪种,其肌内脂肪含量为(1.51±0.19)%[20],因此本研究利用RNA-Seq技术对莱芜猪和大白猪肌内脂肪中circRNA进行鉴定分析,以找出调控猪脂肪沉积的circRNA,进一步了解circRNA调控脂肪沉积和脂代谢作用的机制,为猪肉品质的提高奠定理论基础。
1 材料与方法 1.1 试验动物和样品制备本试验以脂质沉积存在显著差异的大白猪和地方品种莱芜猪为材料,均饲养于莱芜市大千农牧有限公司,在相同饲养条件和环境下生长育肥,参照满足当前营养需要标准(National Research Council, NRC, 1998)饲喂日粮,选择处于屠宰期(约180日龄),健康无病、体况良好,种内个体体重相近的雌性大白猪(约100 kg)和莱芜猪(约35 kg)各3头,采集其背最长肌肌内脂肪组织,参照之前报道的肌内脂肪剥离方法[21],用消毒的眼科镊和手术刀,小心地分离出肌束间的脂肪,以避免肌肉纤维的污染。并且为减少RNA的降解,该过程在冰上进行。试验设置大白猪肌内脂肪组织和莱芜猪肌内脂肪组织两组样本,每组样本3个重复,分别为D-JN-1、D-JN-2、D-JN-3和L-JN-1、L-JN-2、L-JN-3。
1.2 方法 1.2.1 RNA的提取及质控取等量低温保存的脂肪组织样本,根据说明书,利用mirVanaTM RNA提取试剂盒(#AM1561, Ambion, USA)提取每个脂肪组织样本的总RNA,将分离的总RNA样本置于-80 ℃保存。用NanoDrop 2000 (Thermo Scientific, USA)分光光度计测定OD260 nm/OD280 nm值以及RNA样品浓度,并控制在1.9~2.1之间。Bioanalyzer 2100(Agilent, Santa Clara, CA)用于评估总RNA质量,检测RNA完整性(RNA integrity number, RIN),并控制RIN≥7且28S/18S≥0.7。
1.2.2 cDNA文库构建和RNA测序根据Illumina® TruSeqTM RNA样品制备流程,构建6个cDNA文库(包括D-JN-1、D-JN-2、D-JN-3、L-JN-1、L-JN-2和L-JN-3)。构建cDNA文库的步骤主要包括Oligo(dT)磁珠分离富集纯化mRNA,酶促RNA片段化,cDNA的合成,测序接头衔接及PCR扩增等。应用Agilent DNA 1000试剂盒和Agilent 2100生物分析仪(Agilent technologise, Santa Clara, CA)测定cDNA文库大小和纯度。应用ABI stepOnePlus实时荧光定量PCR系统对cDNA文库有效浓度进行准确定量(文库有效浓度>2 nmol·L-1),以保证文库质量。文库质检合格后,利用IlluminaHiSeqTM2500,采用双端测序,对每个cDNA文库进行测序,得到原始测序数据raw reads。
1.2.3 circRNA测序分析使用fastx_toolkit[22]软件(v0.0.14)对raw reads进行质控,去低质量序列、去接头污染、去除rRNA等得到clean reads。采用BWA中的BWA-MEM算法[23]将clean reads比对到参考基因组上,用软件CIRI[24]对circRNA进行识别,对比对结果SAM文件进行扫描,寻找PCC(paired chiastic clipping)和PEM(paired-end mapping)位点,以及GT-AG剪接信号,最后将具有junction位点的序列用动态规划算法进行重新比对,来确保识别circRNA的可靠性。采用DEseq2[25]进行circRNA差异表达分析,比较处理组与参考组,并选取|Fold Change| ≥1.5和P < 0.05作为差异表达阈值,鉴定差异表达的circRNA。
1.2.4 差异表达circRNA的靶基因预测运用miRanda和Targetscan对差异表达的circRNA进行靶标关系预测[26-27],由于circRNA的miRNA结合位点较多,我们在差异表达circRNAs中选出了能较多结合miRNA的circRNAs之后,从Ensemebel数据库中提取了猪所有基因的3′UTR序列,在Linux系统下利用miRanda[26](http://www.microrna.org/microrna)、RNAhybrid[28](http://bibiserv.techfak.uni-bielefeld.de/rnahybrid)、PITA[29](http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html)3种软件对这些miRNAs进行靶基因预测,并取三者预测结果的交集用于后续进一步分析。
1.2.5 circRNA-miRNA-mRNA互作网络图结合本实验室前期所做的莱芜猪与大白猪肌内脂肪组织中差异表达mRNA的鉴定与分析工作[30],筛选出与差异表达circRNA相关的差异表达基因,利用Cytoscape软件对circRNA-miRNA-mRNA构建互作网络图[31]。
1.2.6 靶基因的功能富集分析应用ClueGO[32]软件对与差异表达circRNA相关的差异表达基因进行富集分析。基因本体论(gene ontology, GO, http://www.geneontology.org/)是基因功能国际分类标准,由分子功能(molecular function)、生物过程(biological process)和细胞组分(cellular component)组成。通路富集分析能确定差异表达基因参与的主要代谢途径和信号通路,KEGG (kyoto encyclopedia of genes and genomes)数据库[33](http://www.genome.jp/kegg)作为相关的主要公共数据库,是进行生物体代谢分析、代谢网络研究的强有力工具。基于超几何分布,计算差异表达基因显著富集的GO条目和信号通路,利用Benjamini-Hochberg法对P_value进行校正,当Q_value (corrected P_value)≤0.05时,则显著富集。
1.2.7 实时荧光定量PCR验证qRT-PCR方法用于验证基因的表达水平。取RNA样品反转录合成cDNA模板。利用QuantiFast® SYBR® Green PCR Kit (Qiagen, Germany)和LightCycler® 480 ⅡReal-time PCR Instrument (Roche, Swiss)进行qRT-PCR分析。对4个circRNAs进行qRT-PCR的验证,4个circRNAs对应的引物序列见表 1。其反应体系:1 μL cDNA,0.5 μL正向引物,0.5 μL反向引物,SYBR mix 10 μL,ddH2O 10 μL。反应条件:94 ℃预变性10 min;然后45个循环:94 ℃变性15 s、60 ℃ 60 s、72 ℃ 10 min。以甘油醛-3-磷酸脱氢酶(glyceraldehyde-3-phosphate dehydrogenase, GAPDH)基因为内参,应用2-△△Ct法计算样本间circRNA的相对表达量,用t-检验对相对表达量进行统计分析,数据表示为“平均数±标准差(Mean±SD)”,P < 0.05表示差异显著。
|
|
表 1 基因以及对应的引物 Table 1 Gene and the corresponding primer sequences |
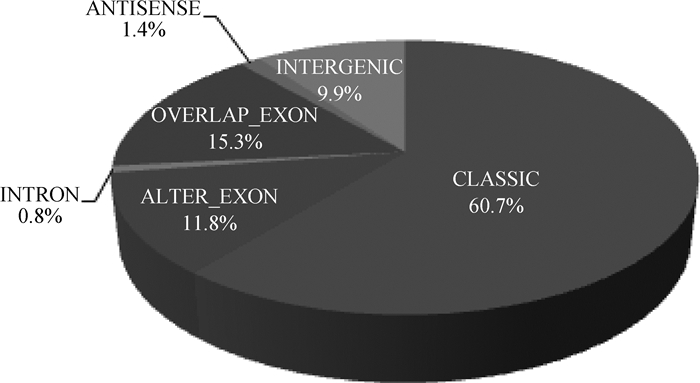
通过环状RNA测序分析,共检测得到9 649个circRNAs,包括CLASSIC、ALTER_EXON、INTRON、OVERLAP_EXON、ANTISENSE和INTERGENIC 6种类型,所占比例如图 1所示。大白猪的3个肌内脂肪组织分别得到88 890 352、89 083 366、91 659 768条reads,比对到总读段数的reads为88 358 935、88 674 923、91 184 863条;莱芜猪的3个肌内脂肪组织分别得到90 180 707、90 392 480、91 093 142条reads,比对到总读段数的reads为89 444 077、89 871 730、90 524 685条,两品种猪比对率均达到95%以上(表 2)。这些结果表明,测序数据质量可靠,为下游数据分析的可靠性奠定了坚实的基础。
|
图 1 各类型circRNAs占总circRNAs的比例 Figure 1 The proportion of various circRNAs in total circRNAs |
|
|
表 2 6个样本比对参考基因组结果 Table 2 The result of 6 samples after mapping to the reference genome |
以|Fold Change| ≥1.5和P < 0.05作为差异表达阈值,筛选得到181个差异表达circRNAs,其中102个上调circRNAs(L vs D),79个下调circRNAs(L vs D)。由于ssc_circ_0002807(下调)、ssc_circ_0009352(上调)有较多miRNA结合位点,因此对其进行后续分析。
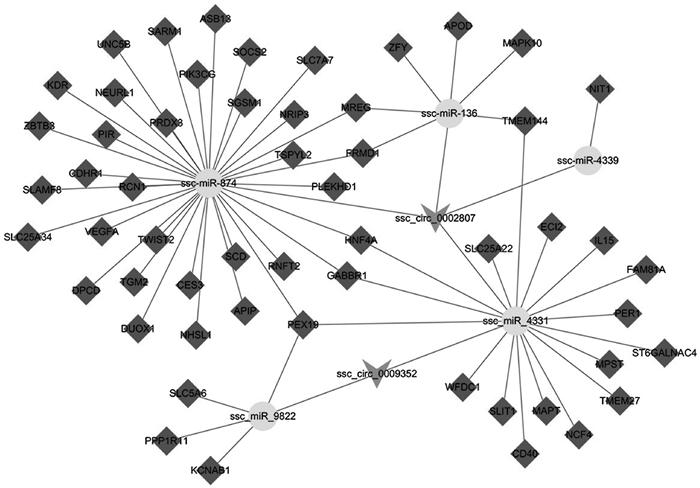
2.3 miRNA结合位点预测及circRNA-miRNA-mRNA互作网络图对具有较多miRNA结合位点的差异表达circRNAs:ssc_circ_0002807和ssc_circ_0009352进行miRNA结合位点的预测(图 2),每四行作为一个靶向关系展示。其中自由能越小,打分值越高,该靶向关系越可靠。ssc-miR-136、ssc-miR-4331、ssc-miR-4339和ssc-miR-874可以结合ssc_circ_0002807,ssc-miR-4331和ssc-miR-9822-3p可以结合ssc_circ_0009352,对ssc_circ_0002807、ssc_circ_0009352及其靶基因构建了互作网络图(图 3)。

|
第1行每个字段分别表示miRNA名称、circRNA id、打分值、最小自由能、miRNA与circRNA基因结合的起始位点、miRNA与circRNA基因的结合终止位点、circRNA基因与miRNA结合的起始位点、circRNA基因与miRNA结合的终止位点;第2行(sRNA)表示反向互补的miRNA序列;第3行表示miRNA每个碱基和circRNA的互补配对情况,竖线表示AT或GC匹配,冒号表示GT匹配;第4行(Ref)表示miRNA结合的circRNA区域 The first line represent miRNA names, circRNA id, score, minimum free energy, starting sites of miRNA combined with circRNA, ending sites of miRNA combined with circRNA, starting sites of circRNA combined with miRNA, ending sites of circRNA combined with miRNA, respectively; The second line (sRNA) represent the reverse complementary miRNA sequence; The third line represent complementary base-pairing of miRNA and circRNA: the vertical indicate matching of A and T or G and C, the colon represent matching of G and T; The fourth line (Ref) represent the circRNA region combined with miRNA 图 2 circRNA与miRNA靶标关系示意图 Figure 2 Schematic diagram of the relationship between circRNA and miRNA |
|
箭头表示circRNAs,圆形表示miRNAs,菱形表示miRNA靶基因 The arrows represent circRNAs, the circles represent miRNAs, the rhomboids represent the target genes of miRNAs 图 3 生物信息学预测的circRNA-miRNA-gene互作网络图 Figure 3 Predicted circRNA-miRNA-gene network for two selected circRNAs by bioinformatics |
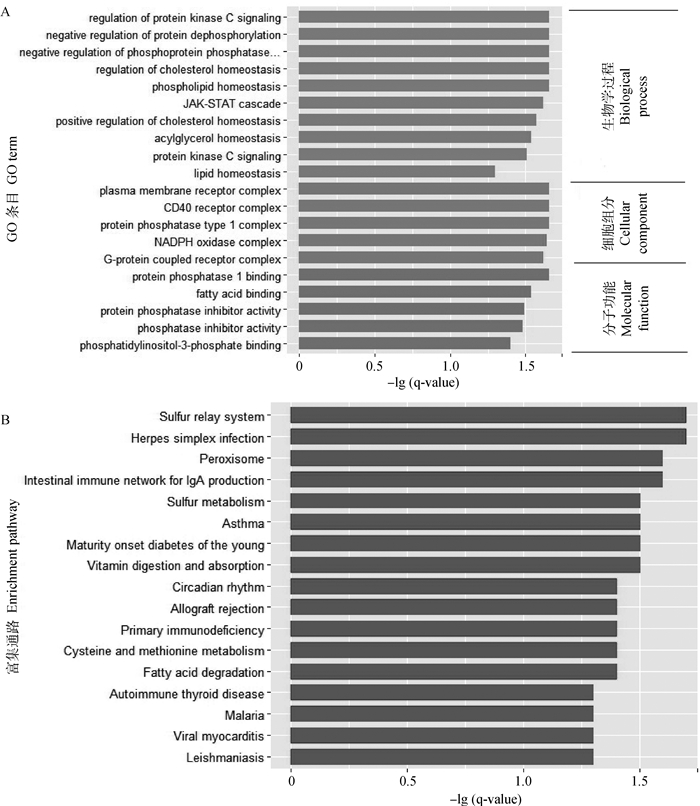
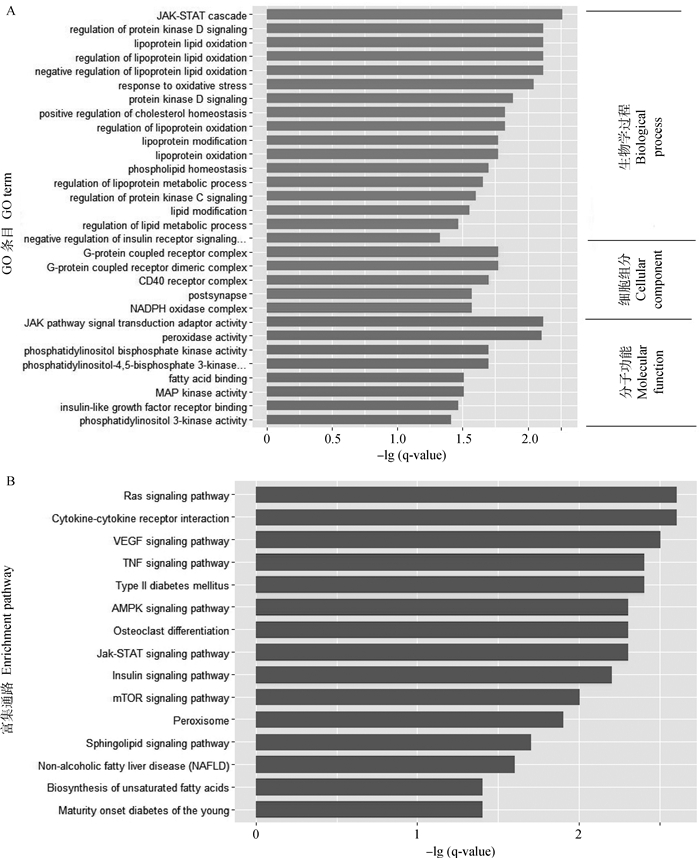
为了深入探究这些靶基因的功能,运用ClueGO进行GO和KEGG富集分析。在生物学过程,发现ssc_circ_0009352的靶基因主要富集于蛋白激酶C活性、蛋白去磷酸化、胆固醇平衡、磷脂平衡、甘油酯平衡和脂质代谢平衡等;在分子生物功能,富集于蛋白磷酸酶、脂肪酸结合、磷脂酰肌醇磷酸结合(图 4A)。而在通路富集中也富集到与脂代谢相关的通路,如过氧化物酶体、脂肪酸降解(图 4B)。对于ssc_circ_0002807,在生物学过程,靶基因中差异表达基因富集于JAK-STAT级联、蛋白激酶D信号调控、脂蛋白脂质氧化、氧化应激反应、胆固醇平衡、脂蛋白修饰及氧化、磷脂平衡、脂质代谢过程等。在分子功能,富集于JAK通道信号转导适配器活动、过氧化物酶活性、脂肪酸结合、MAP激酶活性等(图 5A)。通路富集分析显示,Ras信号通路、细胞因子受体相互作用、血管内皮细胞生长因子信号通路、二型糖尿病、AMPK信号通路、不饱和脂肪酸的生物合成等通路显著富集(图 5B)。
|
图 4 ssc_circ_0009352靶基因GO和KEGG pathway富集分析 Figure 4 GO(A)and KEGG pathway (B) enrichment analysis of the ssc_circ_0009352 targeted genes |
|
图 5 ssc_circ_0002807靶基因GO和KEGG pathway富集分析 Figure 5 GO(A)and KEGG pathway (B) enrichment analysis of the ssc_circ_0002807 targeted genes |
|
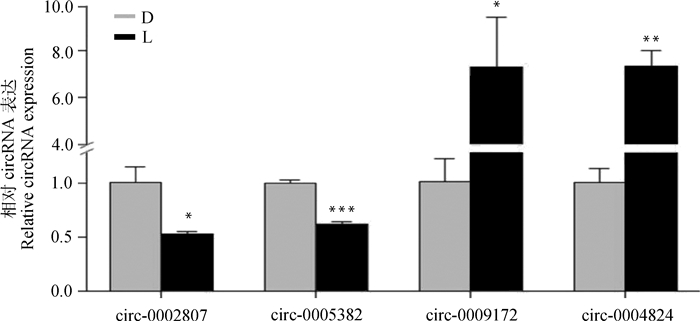
不同猪种间,*. P < 0.05;**. P < 0.01;***. P < 0.001 *. P < 0.05;**. P < 0.01;***. P < 0.001 between different pig breeds 图 6 大白和莱芜肌内脂肪组织间4个差异表达circRNAs的qRT-PCR验证 Figure 6 qRT-PCR validation for the expression of 4 selected differentially expressed circRNAs in intramuscular adipose tissue between Large White and Laiwu pigs |
选择4个差异表达circRNAs进行验证,结果显示,在莱芜肌内脂肪组织中ssc_circ_0002807(chr14:52135828..52247318:-)、ssc_circ_0005382(chr1:216338622..216447657:-)显著下调;ssc_circ_0009172(chr6:148414520..148422837:+)、ssc_circ_0004824(chr1:120698326..120698912:-)显著上调,与测序结果完全一致。
3 讨论本研究对大白猪和莱芜猪肌内脂肪组织中的circRNA进行鉴定和分析,共鉴定到9 649个circRNAs,有181个circRNAs差异表达,其中102个circRNAs在莱芜猪肌内脂肪组织中上调表达,79个下调。通过qRT-PCR,验证了ssc_circ_0002807、ssc_circ_0005382、ssc_circ_0009172和ssc_circ_0004824,与测序结果一致。circRNA是一类稳定的非编码RNA,广泛存在于不同的细胞中,在基因的表达中发挥了关键的调节作用[34]。circRNA具有高度稳定性,多样性,丰富的miRNA反应元件(miRNA Response Elements, MRE),序列特异性和物种间保守性[35],这些特征表明,circRNA在转录及转录后水平发挥重要作用,可能参与调控脂质沉积,为代谢相关性疾病提供潜在的预防和治疗靶标。
最近研究表明,在许多真核细胞中circRNA可以作为miRNA海绵来调控基因表达。Hansen等[12-13]报道,ciRs7在人脑组织中丰富表达,它含有多个串联的miR-7结合位点,可以作为内源性的miRNA海绵吸附miR-7,在正常生理条件下ciRs7-miR-7-miR-7的靶基因处于动态平衡,当这种稳态被打破时则可能诱发疾病。本研究进行了circRNA-miRNA靶标关系预测。这些miRNA中,miR-136作为ssc_circ_0002807的靶向,与ASCL1、PPARα、C/EBPα呈负相关。而在脂肪组织,PPARα和PPARγ可以诱导ASCL1基因,从而酯化长链脂肪酸[36-38]。miR-4331作为ssc_circ_0002807和ssc_circ_0009352的靶向,能够激活p38MAPK通路[39],激活的p38MAPK可以促进骨髓间充质干细胞的脂肪形成[40]。由此推测,ssc_circ_0002807可能参与成脂分化的调控。还有报道指出在感染HIV患者的脂肪组织中,一些miRNA异常表达,其中miR-874上调表达,这些miRNA的表达量变化可能导致与HIV相关的脂肪代谢障碍[41],而miR-874是ssc_circ_0002807的靶向,由此推测,ssc_circ_0002807可能参与脂质代谢的调控。
竞争性内源RNA(competing endogenous RNAs, ceRNA)是通过miRNA反应元件竞争结合共同的miRNA来实现相互调控作用的转录产物,该机制也就是所谓的“ceRNA”假说。根据这一理论,circRNA和mRNA可以结合相同的miRNA[42],为了深入探索circRNA在脂肪沉积和代谢方面的功能,我们构建了circRNA-miRNA-gene互作网络图(图 3)。通过GO和KEGG富集分析发现,ssc_circ_0009352和ssc_circ_0002807的靶基因富集于与脂类代谢相关的生物学过程,如氧化应激反应、胆固醇平衡、磷脂平衡、脂质代谢过程等。由此推断,ssc_circ_0009352、ssc_circ_0002807可能在脂肪代谢中发挥作用。另外,ssc_circ_0002807的靶基因通路中还富集到二型糖尿病、胰岛素信号通路和非酒精性脂肪肝等。脂肪组织能够储存大量的甘油三酯和脂溶性物质以及机体代谢产生的多余能量,如果脂肪组织吸收和储存循环脂质的能力失调,则可能会导致其它非脂肪组织中脂质的积累,导致肥胖和能量代谢平衡失调,引发肥胖相关性疾病的产生,如二型糖尿病、动脉粥样硬化、胰岛素抵抗、非酒精性脂肪肝炎、心血管疾病等[43-44]。生物信息学分析显示,circRNA可能参与调控脂肪沉积和脂质代谢,尽管如此,关于circRNA在脂肪沉积和脂质代谢的功能以及circRNA-miRNA-gene的互作关系还需要进一步探索和验证。
4 结论本研究鉴定分析了大白和莱芜猪肌内脂肪组织中circRNA的表达情况,发现181个差异表达circRNAs。对具有较多miRNA结合位点的ssc_circ_0002807和ssc_circ_0009352靶基因进行了GO和KEGG富集分析,发现靶基因富集于胆固醇平衡和磷脂平衡等与脂代谢相关的生物学过程。并通过qRT-PCR验证了ssc_circ_0002807、ssc_circ_0005382、ssc_circ_0009172和ssc_circ_0004824,与测序结果一致,推测ssc_circ_0002807、ssc_circ_0009352可能参与调控脂肪沉积和脂质代谢。
| [1] |
王蜀金, 郭春华. 全球猪肉市场纵览:附加生产力刺激世界猪肉产量[J]. 猪业科学, 2014(12): 24–25.
WANG S J, GUO C H. Global pork market overview:additional productivity stimulates world pork production[J]. Swine Industry Science, 2014(12): 24–25. DOI: 10.3969/j.issn.1673-5358.2014.12.006 (in Chinese) |
| [2] | DODSON M V, JIANG Z H, CHEN J, et al. Allied industry approaches to alter intramuscular fat content and composition in beef animals[J]. J Food Sci, 2010, 75(1): R1–R8. DOI: 10.1111/jfds.2010.75.issue-1 |
| [3] | LISTRAT A, LEBRET B, LOUVEAU I, et al. How muscle structure and composition influence meat and flesh quality[J]. Sci World J, 2016, 2016: 3182746. |
| [4] |
尹靖东, 李德发. 猪肉质形成的分子机制与营养调控[J]. 动物营养学报, 2014, 26(10): 2979–2985.
YIN J D, LI D F. Molecular mechanism underlying meat quality formation and its nutritional regulation in pigs[J]. Chinese Journal of Animal Nutrition, 2014, 26(10): 2979–2985. DOI: 10.3969/j.issn.1006-267x.2014.10.009 (in Chinese) |
| [5] | LUKIW W J. Circular RNA (circRNA) in Alzheimer's disease (AD)[J]. Front Genet, 2013, 4: 307. |
| [6] | BACHMAYR-HEYDA A, REINER A T, AUER K, et al. Correlation of circular RNA abundance with proliferation-exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues[J]. Sci Rep, 2015, 5: 8057. DOI: 10.1038/srep08057 |
| [7] | QIN M L, LIU G, HUO X S, et al. Hsa_circ_0001649:A circular RNA and potential novel biomarker for hepatocellular carcinoma[J]. Cancer Biomark, 2016, 16(1): 161–169. DOI: 10.3233/CBM-150552 |
| [8] |
姜隽, 杨悠, 姜睿, 等. 环状RNA对基因的调控作用及其与心血管疾病的关系[J]. 中华心血管病杂志, 2016, 44(4): 364–366.
JIANG J, YANG Y, JIANG R, et al. Regulating mechanisms of circRNA and their relationship with cardiovascular diseases[J]. Chinese Journal of Cardiology, 2016, 44(4): 364–366. (in Chinese) |
| [9] | LU D, XU A D. Mini review:circular RNAs as potential clinical biomarkers for disorders in the central nervous system[J]. Front Genet, 2016, 7: 53. |
| [10] | XIE H J, REN X L, XIN S N, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer[J]. Oncotarget, 2016, 7(18): 26680–26691. |
| [11] | DANG Y J, YAN L Y, HU B Q, et al. Tracing the expression of circular RNAs in human pre-implantation embryos[J]. Genome Biol, 2016, 17: 130. DOI: 10.1186/s13059-016-0991-3 |
| [12] | HANSEN T B, JENSEN T I, CLAUSEN B H, et al. Natural RNA circles function as efficient microRNA sponges[J]. Nature, 2013, 495(7441): 384–388. DOI: 10.1038/nature11993 |
| [13] | HANSEN T B, KJEMS J, DAMGAARD C K. Circular RNA and miR-7 in cancer[J]. Cancer Res, 2013, 73(18): 5609–5612. DOI: 10.1158/0008-5472.CAN-13-1568 |
| [14] |
李美航, 仇杨, 刘帅, 等. 过表达miR-103促进猪前体脂肪细胞分化[J]. 生物工程学报, 2012, 28(8): 927–936.
LI M H, QIU Y, LIU S, et al. Over-expressed miR-103 promotes porcine adipocyte differentiation[J]. Chinese Journal of Biotechnology, 2012, 28(8): 927–936. (in Chinese) |
| [15] | LI H X, ZHANG Z, ZHOU X, et al. Effects of microRNA-143 in the differentiation and proliferation of bovine intramuscular preadipocytes[J]. Mol Biol Rep, 2010, 38(7): 4273–4280. |
| [16] | MIGDAL L, KOZIOL K, PALKA S, et al. Mutations in leptin (LEP) gene are associated with carcass and meat quality traits in crossbreed rabbits[J]. Anim Biotechnol, 2017, 29(2): 153–159. |
| [17] | SERĀO N V, VERONEZE R, RIBEIRO A M, et al. Candidate gene expression and intramuscular fat content in pigs[J]. J Anim Breed Genet, 2011, 128(1): 28–34. DOI: 10.1111/j.1439-0388.2010.00887.x |
| [18] | WON S, JUNG J, PARK E, et al. Identification of genes related to intramuscular fat content of pigs using genome-wide association study[J]. Asian-Australas J Anim Sci, 2018, 31(2): 157–162. DOI: 10.5713/ajas.17.0218 |
| [19] |
曾勇庆, 王根林, 魏述东, 等. 含不同比例莱芜猪血缘杂交猪胴体品质及肉质特性的研究[J]. 遗传, 2005, 27(1): 65–69.
ZENG Q Y, WANG G L, WEI S D, et al. Studies on carcass and meat quality performance of crossbred pigs with graded proportions of Laiwu Black genes[J]. Hereditas, 2005, 27(1): 65–69. (in Chinese) |
| [20] |
于永生, 张志彬, 李娜, 等. 不同日龄东北民猪与大白猪背最长肌游离水和肌内脂肪含量的测定[J]. 养猪, 2015(1): 47–48.
YU Y S, ZHANG Z B, LI N, et al. Determination of the maximum muscle free water and intramuscular fat content of Northeast China pigs and big white pigs with different age[J]. Swine Production, 2015(1): 47–48. (in Chinese) |
| [21] | SUN W X, WANG H H, JIANG B C, et al. Global comparison of gene expression between subcutaneous and intramuscular adipose tissue of mature Erhualian pig[J]. Genet Mol Res, 2013, 12(4): 5085–5101. DOI: 10.4238/2013.October.29.3 |
| [22] | GARDON A, HANNON G J. FASTX-TOOLKIT. FASTQ/A short-reads pre-processing tools[EB/OL]. (2010). http://hannonlab.cshl.edu/fastx_toolkit. |
| [23] | LI H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM[C]. arXiv preprint arXiv: 1303. 3997, 2013. |
| [24] | GAO Y, WANG J F, ZHAO F Q. CIRI:an efficient and unbiased algorithm for de novo circular RNA identification[J]. Genome Biol, 2015, 16: 4. DOI: 10.1186/s13059-014-0571-3 |
| [25] | LOVE M I, HUBER W, ANDERS S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2[J]. Genome Biol, 2014, 15(12): 550. DOI: 10.1186/s13059-014-0550-8 |
| [26] | ENRIGHT A J, JOHN B, GAUL U, et al. microRNA targets in Drosophila[J]. Genome Biol, 2003, 5(1): R1. |
| [27] | PASQUINELLI A E. microRNAs and their targets:recognition, regulation and an emerging reciprocal relationship[J]. Nat Rev Genet, 2012, 13(4): 271–282. DOI: 10.1038/nrg3162 |
| [28] | REHMSMEIER M, STEFFEN P, HÖECHSMANN M, et al. Fast and effective prediction of microRNA/target duplexes[J]. RNA, 2004, 10(10): 1507–1517. DOI: 10.1261/rna.5248604 |
| [29] | KERTESZ M, IOVINO N, UNNERSTALL U, et al. The role of site accessibility in microRNA target recognition[J]. Nat Genet, 2007, 39(10): 1278–1284. DOI: 10.1038/ng2135 |
| [30] |
黄万龙. 两品种猪不同脂肪组织中长链非编码RNA的鉴定及其功能分析[D]. 北京: 中国农业科学院, 2017.
HUANG W L. Identification and functional analysis of long non-coding RNA in distinct adipose tissues from two pig breeds[D]. Beijing: Chinese Academy of Agricultural Sciences, 2017. (in Chinese) |
| [31] | SMOOT M E, ONO K, RUSCHEINSKI J, et al. Cytoscape 2.8:new features for data integration and network visualization[J]. Bioinformatics, 2011, 27(3): 431–432. |
| [32] | BINDEA G, MLECNIK B, HACKL H, et al. ClueGO:a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks[J]. Bioinformatics, 2009, 25(8): 1091–1093. DOI: 10.1093/bioinformatics/btp101 |
| [33] | KANEHISA M, ARAKI M, GOTO S, et al. KEGG for linking genomes to life and the environment[J]. Nucleic Acids Res, 2008, 36(Database issue): D480–D484. |
| [34] | JECK W R, SORRENTINO J A, WANG K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats[J]. RNA, 2013, 19(2): 141–157. DOI: 10.1261/rna.035667.112 |
| [35] | CORTÉS-LÓPEZ M, MIURA P. Emerging functions of circular RNAs[J]. Yale J Biol Med, 2016, 89(4): 527–537. |
| [36] | WANG N D, FINEGOLD M J, BRADLEY A, et al. Impaired energy homeostasis in C/EBPα knockout mice[J]. Science, 1995, 269(5227): 1108–1112. DOI: 10.1126/science.7652557 |
| [37] | LOBO S, WICZER B M, BERNLOHR D A. Functional analysis of long-chain acyl-CoA synthetase 1 in 3T3-L1 adipocytes[J]. J Biol Chem, 2009, 284(27): 18347–18356. DOI: 10.1074/jbc.M109.017244 |
| [38] | RUBINOW K B, WALL V Z, NELSON J, et al. Acyl-CoA synthetase 1 is induced by gram-negative bacteria and lipopolysaccharide and is required for phospholipid turnover in stimulated macrophages[J]. J Biol Chem, 2013, 288(14): 9957–9970. DOI: 10.1074/jbc.M113.458372 |
| [39] | ZHAO X M, BAI X Y, GUAN L Y, et al. microRNA-4331 promotes TGEV-induced mitochondrial damage via targeting RB1, up-regulating IL1RAP, and activating p38 MAPK pathway in vitro[J]. Mol Cell Proteomics, 2018, 17(2): 190–204. DOI: 10.1074/mcp.RA117.000432 |
| [40] | ZHAO Q, LU Y Z, YU H Y, et al. Low magnitude high frequency vibration promotes adipogenic differentiation of bone marrow stem cells via P38 MAPK signal[J]. PLoS One, 2017, 12(3): e0172954. DOI: 10.1371/journal.pone.0172954 |
| [41] | SQUILLACE N, BRESCIANI E, TORSELLO A, et al. Changes in subcutaneous adipose tissue microRNA expression in HIV-infected patients[J]. J Antimicrob Chemother, 2014, 69(11): 3067–3075. DOI: 10.1093/jac/dku264 |
| [42] | CHEN Y H, LI C, TAN C L, et al. Circular RNAs:a new frontier in the study of human diseases[J]. J Med Genet, 2016, 53(6): 359–365. DOI: 10.1136/jmedgenet-2016-103758 |
| [43] | WANG S X, SONG K X, SRIVASTAVA R, et al. Nonalcoholic fatty liver disease induced by noncanonical Wnt and its rescue by Wnt3a[J]. FASEB J, 2015, 29(8): 3436–3445. DOI: 10.1096/fj.15-271171 |
| [44] | KLÖTING N, BLVHER M. Adipocyte dysfunction, inflammation and metabolic syndrome[J]. Rev Endocr Metab Disord, 2014, 15(4): 277–287. DOI: 10.1007/s11154-014-9301-0 |



