2. 福建光华百斯特生态农牧发展有限公司, 三明 365106
2. Fujian Guanghua BEST Eco-agriculture and Animal Husbandry Development Co. Ltd., Sanming 365106, China
Nhlh2基因也称为NSCL-2,该基因编码的蛋白属于碱性螺旋-环-螺旋转录因子,由V.Göbel等[1]在1991年首次在小鼠胎脑cDNA文库中筛选分离得到。研究表明,Nhlh2通过调控下丘脑GnRH的分泌,参与调控小鼠初情期的启动[2-3]。与正常小鼠相比,Nhlh2突变导致下丘脑GnRH神经元数量减少50%,同时体内GnRH的浓度下降80%~90%[3-4]。Nhlh2敲除的小鼠在胚胎发育初期阶段,GnRH神经元滞留在嗅觉/视前区,并表现出非正常性凋亡[5]。Nhlh2敲除的雌性小鼠,大部分卵泡在窦状卵泡阶段停止生长发育[4],排卵数下降50%、发情间期延长及繁殖性能显著下降[6]。此外,D.Nonneman等[7]通过全基因组关联分析发现,与母猪初情期启动失败高度关联的SNP位点位于Nhlh2基因附近。这些结果表明,Nhlh2可能参与调控哺乳动物卵泡的成熟、排卵及初情期启动等繁殖活动。但是目前关于Nhlh2基因的表达调控机制仍不清楚。
本研究以猪卵巢基因组DNA为模板,以猪颗粒细胞为细胞模型,通过克隆猪Nhlh2基因的启动子区域序列,构建不同缺失长度的报告基因系统,分析了该基因的启动子活性,并研究了部分转录因子与该基因的表达调控关系。本研究能够为后续母猪Nhlh2基因的结构和功能研究提供分子基础,也为母猪繁殖机制的研究提供一定的参考。
1 材料与方法试验于2016年5月至2017年4月在华南农业大学广东省农业动物基因组学与分子育种重点实验室完成。
1.1 试验材料试验动物及相关组织:用于猪Nhlh2启动子克隆的DNA模板样品来自中山某猪场的长大二元杂交母猪的卵巢组织,猪卵巢颗粒细胞是用广州市某屠宰场商品猪卵巢组织培养的原代细胞。
分子克隆试验材料:基因组DNA抽提试剂、无内毒素质粒小量提取试剂、普通质粒小量提取试剂、凝胶回收试剂购自美国Magen公司,pMD-18T Vector、DNA A-Tailing Kit、PrimeStar HS、T4连接酶、限制性内切酶、10×Loading Buffer购自美国TaKaRa公司,Trans5α Chemically Competent Cell购自北京全式金有限公司,YY1 Antibody、C/EBPβ Antibody、GAPDH Antibody购自上海absin有限公司。
细胞培养与检测试剂:培养基、青链霉素(双抗溶液)购自Invitrogen公司,胎牛血清、胰蛋白酶购自GIBCO,用于细胞转染的Lipofectamine@ 3000购自美国Invitrogen公司,YY1-siRNA和C/EBPβ-siRNA由广州锐博生物有限公司合成,pGL3-basic载体、pCDNA3.1载体、荧光素酶报告基因检测试剂盒购自Promega公司。其他均为常规化学试剂。
1.2 试验方法 1.2.1 猪Nhlh2启动子的克隆根据NCBI基因数据库中猪Nhlh2基因5′上游序列,设计合适的引物(表 1),使用PrimeStar HS高保真酶,以猪卵巢组织DNA为模板,PCR克隆扩增猪Nhlh2基因5′上游-3 565~+129 bp片段。PCR采用10 μL反应体系:5 μL PrimeSTAR HS,0.3 μmol·L-1上下游引物,0.3 μL DNA模板,ddH2O补足至10 μL。PCR反应条件:98 ℃变性10 s,60 ℃退火15 s,72 ℃延伸4 min,共循环35次。PCR产物经1.5%琼脂糖凝胶电泳后回收加A尾,取4 μL产物加1 μL pMD18-T载体,16 ℃连接反应5 h,转化大肠埃希菌DH5α,培养12 h,挑取单个菌落、扩增、测序。测序鉴定正确的质粒命名为T-Nhlh2。
|
|
表 1 引物列表 Table 1 Primers used in this study |
以测序鉴定正确的T-Nhlh2为模板,设计8条不同的上游引物,PCR扩增8条不同5′端的缺失片段(扩增引物见表 1),琼脂糖凝胶电泳回收目的片段,用Mlu Ⅰ、Xho Ⅰ双酶切胶回收系列缺失片段PCR产物,同时pGL3-basic经Mlu Ⅰ、Xho Ⅰ双酶切,定向连接,转化大肠埃希菌DH5α,氨苄青霉素平板涂板后,培养过夜进行抗性筛选,阳性克隆经双酶切鉴定正确后,命名为P1~P8。利用TFBIND[8]和JASPAR[9]等生物信息学网站预测分析目的片段存在的潜在转录因子结合位点。
1.2.3 转录因子YY1、C/EBPβ表达载体、突变体的构建根据YY1、C/EBPβ基因的序列,分别设计引物(表 1),反应条件及体系按PrimeStar HS PCR说明书进行,YY1经Nhe Ⅰ、Xho Ⅰ双酶切,C/EBPβ经Kpn Ⅰ、Xho Ⅰ双酶切,分别连接pCDNA3.1载体获得pCDNA3.1-YY1、pCDNA3.1-C/EBPβ。以P7(-238~+129)为模板,根据重叠PCR原理,设计含有突变碱基的引物(表 1),构建YY1、C/EBPβ潜在结合位点突变的突变载体,命名为pGL3-P7-YY1-Mut、pGL3-P7-C/EBPβ-Mut。
1.2.4 猪卵巢颗粒细胞的培养及转染原代颗粒细胞的培养及传代参考文献[10-11],待细胞汇合度约90%时,以2.5×105个·孔-1接种于24孔板,按照Invitrogen公司的Lipofectimine 3000转染试剂盒说明书操作。每个试验设置4个复孔,转染后在无双抗的10%胎牛血清DMEM培养基500 μL中培养6 h后更换为含有双抗的10%胎牛血清DMEM培养基中继续培养,在瞬时转染细胞24 h后,用双荧光素酶报告基因系统进行荧光素酶活性检测,收集数据分析启动子片段转录活性。
1.2.5 染色质免疫共沉淀(ChIP)ChIP试验的操作流程参照文献[12-13],用终浓度为1%甲醛在37 ℃孵育细胞10 min,使蛋白与DNA交联。用终浓度为0.125 mol·L-1的甘氨酸室温处理5 min终止交联。收集细胞后超声破碎。10 000 ×g,4 ℃离心细胞液,收集上清。用特异性抗体与DNA结合蛋白结合,对应抗体为,阳性对照:Anti-RNA Polymerase Ⅱ抗体;阴性对照:兔IgG抗体;试验组:YY1或C/EBPβ特异性抗体,沉淀法分离复合体。反向交联操作释放出DNA,并消化蛋白质。PCR检测Nhlh2启动子-101~-85和-153~-140处片段,引物序列见表 2。
|
|
表 2 ChIP验证引物信息 Table 2 Primers used in ChIP |
所有数据均用“平均值±标准差”表示,并经双尾T-test检验差异是否具有统计学意义。*表示P < 0.05,**表示P < 0.01。
2 结果 2.1 猪Nhlh2启动子缺失片段报告基因重组质粒的构建和酶切鉴定从猪卵巢基因组DNA中克隆获得-3 565~+129(3 694 bp)的Nhlh2的5′端片段(图 1A),并命名为T-Nhlh2。以T-Nhlh2为模板,设计8条不同的上游引物,PCR扩增出8条不同5′端的缺失片段(图 1B),经琼脂糖凝胶电泳分析验证,扩增产物与预期片段大小相符(图 1C)。用限制性内切酶Mlu Ⅰ、Xho Ⅰ酶切这些5′端的缺失片段和双荧光素酶载体后,构建了P1~P8报告基因系统重组质粒,并经双酶切(图 1D)和测序鉴定。
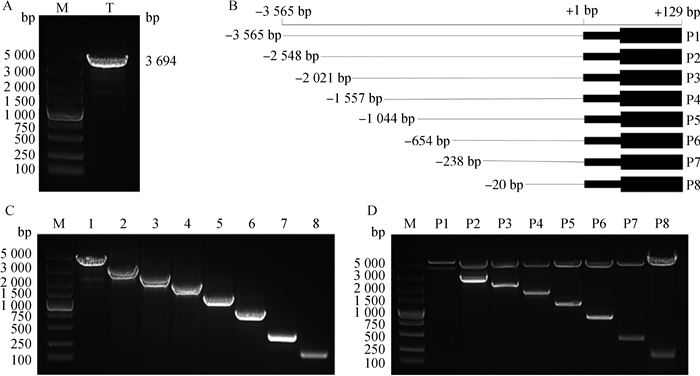
|
A. Nhlh2启动子扩增产物;B. Nhlh2启动子5′端缺失片段长度图示;C. Nhlh2启动子5′端缺失片段扩增产物;D. Nhlh2启动子5′端缺失片段的酶切鉴定。M. DNA相对分子质量标准;T. 3 694 bp(T-Nhlh2);1~8. Nhlh2基因5′端逐段缺失扩增结果(-3 565~+129、-2 548~+129、-2 021~+129、-1 557~+129、-1 044~+129、-654~+129、-238~+129、-20~+129);P1~P8. P1~P8载体的酶切鉴定 A. Products of Nhlh2 promoter amplification; B. Length of Nhlh2 promoter 5′ fragments; C. Products of 5′ fragments of Nhlh2 promoter amplification; D. Identification of Nhlh2 promoter 5′ deletion fragments by restriction enzymes. M. DNA marker DL5000; T. 3 694 bp (T-Nhlh2); 1-8. The DNA bands of 5′ fragments of Nhlh2 gene unidirectional deletions (-3 565-+129, -2 548-+129,-2 021-+129, -1 557-+129, -1 044-+129,-654-+129,-238-+129, -20-+129); P1-P8. Identification of vectors P1-P8 by restriction enzymes 图 1 Nhlh2启动子片段缺失报告载体的构建 Figure 1 Construction of Nhlh2 promoter fragment deletion vectors |
将构建的8个启动子缺失片段的报告基因重组质粒瞬时转染至猪卵巢颗粒细胞,24 h后裂解细胞并收集裂解液进行双荧光素酶活性检测。结果显示,P1~P6片段双荧光素酶活性差异不显著;P7(-238~+129)区域的相对活性最高,说明猪Nhlh2基因的核心启动子区域为-238~+129;与P6(-654~+129)相比,P7片段活性显著升高了1.24倍(P=0.001 8),表明-654~-238片段范围可能存在负调控元件(图 2);与P8(-20~+129)片段相比,P7片段荧光活性下降了9.57倍(P=0.000 1),表明-238~-20区域可能存在正调控元件(图 2)。
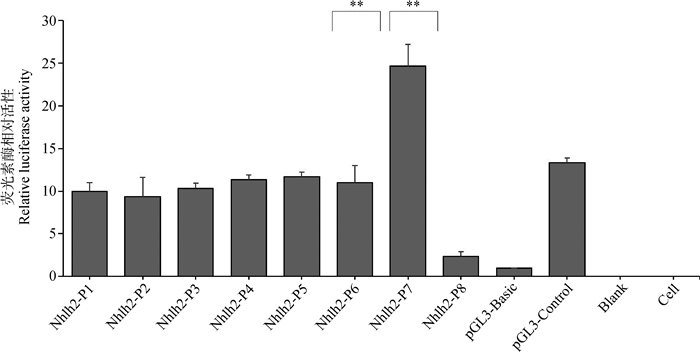
|
**. P<0.01, the same as below 图 2 Nhlh2的5′端缺失片段启动子活性 Figure 2 Luciferase activities of Nhlh2 promoter 5′ deletion fragments |
结合TFBIND、JASPAR等生物信息学软件对-238~-20区域进行预测,发现该区域存在多个潜在的转录因子结合位点,本研究关注YY1和C/EBPβ与Nhlh2之间的表达调控关系。ChIP试验结果表明,转录因子YY1识别并结合在Nhlh2基因-101~-85区域(图 3A);转录因子C/EBPβ识别并结合在Nhlh2基因-153~-140区域(图 3B)。
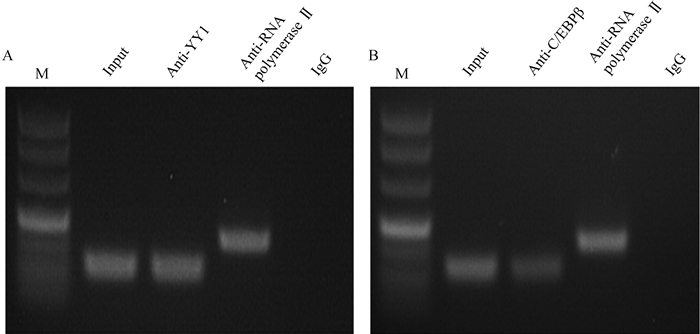
|
A. YY1结合位点ChIP验证结果;B. C/EBPβ结合位点ChIP验证结果。M. DNA相对分子质量标准;Anti-YY1/Anti-C/EBPβ.试验组;input/Anti-RNA polymerase Ⅱ.阳性对照;IgG.阴性对照 A. ChIP validation of YY1 binding site at Nhlh2; B. ChIP validation of C/EBPβ binding site at Nhlh2. M. DNA marker DL500; Anti-YY1/Anti-C/EBPβ. Experimental group; Input/Anti-RNA polymerase Ⅱ. Positive control; IgG. Negative control 图 3 Nhlh2基因启动子区转录因子结合位点ChIP验证 Figure 3 The results of transcription factor binding sites at Nhlh2 gene promoter by ChIP |
如图 4和图 5所示,以pGL3-P7为野生型,构建两个转录因子结合位点的突变体pGL3-P7-YY1-Mut、pGL3-P7-C/EBPβ-Mut,并构建了pCDNA3.1-YY1和pCDNA3.1-C/EBPβ两个真核表达载体及干扰小RNA(YY1-siRNA和C/EBPβ-siRNA)转染至猪卵巢颗粒细胞,探索YY1和C/EBPβ与Nhlh2之间的表达调控关系。
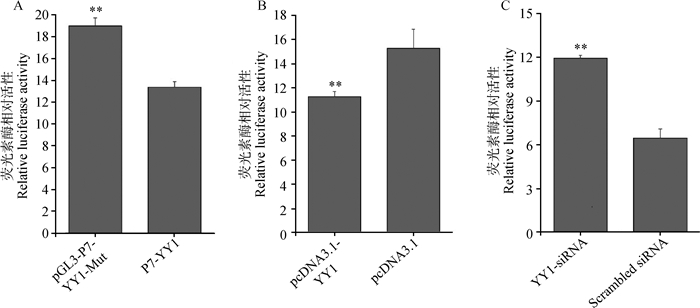
|
A. YY1结合位点突变对Nhlh2启动子活性影响;B.超表达YY1对Nhlh2启动子活性影响;C. YY1-siRNA对Nhlh2启动子活性影响 A. Effects of YY1 binding site mutation on the promoter activity of Nhlh2; B. Effects of overexpressed YY1 on the promoter activity of Nhlh2; C. Effects of YY1-siRNA on the promoter activity of Nhlh2 图 4 YY1对猪Nhlh2启动子活性的影响 Figure 4 Effects of YY1 on the promoter activity of porcine Nhlh2 gene |
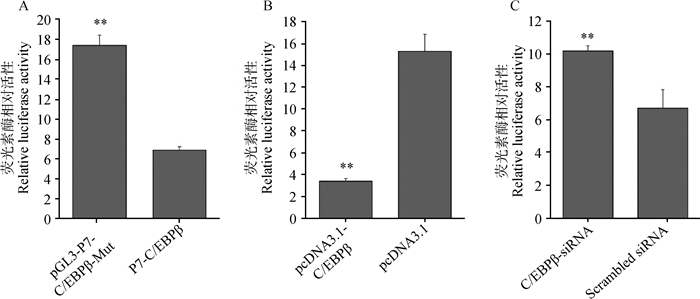
结果显示,pGL3-P7-YY1-Mut与野生型相比,双荧光素酶活性上调0.42倍(P=7.22×10-11)(图 4A);超表达YY1后,pGL3-P7的双荧光素酶活性下调了0.36倍(P=1.28×10-2)(图 4B);抑制YY1的表达后,pGL3-P7的双荧光素酶活性上调了0.85倍(P=2.79×10-4)(图 4C)。pGL3-P7-C/EBPβ-Mut与其野生型相比,双荧光素酶活性上调1.53倍(P=2.23×10-13)(图 5A);超表达C/EBPβ后,pGL3-P7的双荧光素酶活性下调了3.49倍(P=2.03×10-4)(图 5B);抑制C/EBPβ的表达后,pGL3-P7的双荧光素酶活性上调了0.52倍(P=1.35×10-2)(图 5C)。
|
A. C/EBPβ结合位点突变对Nhlh2启动子活性影响;B.超表达C/EBPβ对Nhlh2启动子活性影响;C. C/EBPβ-siRNA对Nhlh2启动子活性影响 A. Effects of C/EBPβ binding site mutation on the promoter activity of Nhlh2; B. Effects of overexpressed C/EBPβ on the promoter activity of Nhlh2; C. Effects of C/EBPβ-siRNA on the promoter activity of Nhlh2 图 5 C/EBPβ对猪Nhlh2启动子活性的影响 Figure 5 Effects of C/EBPβ on the promoter activity of porcine Nhlh2 gene |
大量研究表明,哺乳动物的Nhlh2参与调控卵泡的成熟、排卵及初情期启动等繁殖活动[4, 6],Nhlh2可能是影响母猪繁殖活动的重要候选基因。本研究以长大二元杂交母猪卵巢的基因组DNA为模板,成功构建了8个Nhlh2基因不同缺失长度的启动子双荧光素酶报告载体,双荧光活性检测结果表明,P7的启动子活性最高,说明Nhlh2基因的核心启动子区域为-238~+129(图 2)。同时,P7的启动子活性是P6的2.24倍、P8的10.57倍,表明-654~-238片段范围可能存在负调控元件,而-238~-20区域可能存在正调控元件(图 2)。另外,P8的启动子活性与pGL3-basic空载体相近,因此P8可能无启动子活性。
生物信息学分析表明,Nhlh2启动子区-101~-85和-153~-140区域分别是转录因子YY1、C/EBPβ的潜在结合位点。本试验利用ChIP技术验证了转录因子YY1和C/EBPβ结合在Nhlh2基因核心启动子区域(图 3)。双荧光素酶活性检测结果显示,pGL3-P7-YY1-Mut与野生型相比,双荧光素酶活性显著上调;超表达YY1后,pGL3-P7-YY1的双荧光素酶活性显著下调,抑制YY1的表达后,pGL3-P7-YY1的双荧光素酶活性显著上调。同时,pGL3-P7-C/EBPβ-Mut与野生型相比,双荧光素酶活性显著上调,超表达C/EBPβ后,pGL3-P7-C/EBPβ的双荧光素酶活性显著下调,抑制C/EBPβ的表达后,pGL3-P7-C/EBPβ的双荧光素酶活性显著上调,这些结果表明,转录因子YY1、C/EBPβ结合在Nhlh2基因启动子上抑制其转录活性。
研究表明,转录因子YY1通过结合在Kiss1基因的启动子上,参与并调控下丘脑GnRH的分泌,进而影响雌性动物初情期启动[14-15]。YY1突变雌性个体表现出发情启动失败、无性行为、不育[16],并且在卵母细胞和颗粒细胞之间的信号传导中扮演重要角色,参与调控卵泡颗粒细胞的生长发育与卵母细胞成熟[17-18]。更重要的是,在哺乳动物初情期启动过程中,YY1结合在大量表达上调基因的启动子上[19]。转录因子C/EBPβ是一类结合于DNA增强子区域的多功能反式作用因子,其也和卵巢卵泡的发育密切相关[20-21]。研究表明,特异性敲除卵巢颗粒细胞C/EBPβ后,卵泡发育停滞,无法发生排卵和形成黄体[22]。同时,研究发现,C/EBPβ调控卵巢雌激素和孕酮的合成[23-24],如果抑制卵巢C/EBPβ的表达,将引起小鼠的排卵异常[25],导致小鼠无生育能力[26]。C/EBPβ也可以调节瘦素受体基因的表达,进而调节小鼠排卵[27]。这些研究结果表明,YY1和C/EBPβ参与调控哺乳动物卵泡的成熟、排卵及初情期启动等繁殖活动,也佐证了Nhlh2可能是影响母猪繁殖活动的重要候选基因。
4 结论本研究通过构建猪Nhlh2启动子缺失片段报告基因系统,发现Nhlh2基因的核心启动子区域为-238~+129,-654~-238片段范围可能存在负调控元件,而-238~-20区域可能存在正调控元件;转录因子YY1、C/EBPβ结合在Nhlh2基因启动子上抑制其转录活性。本研究为后续母猪Nhlh2基因的结构和功能研究提供了分子基础。
| [1] | GÖBEL V, LIPKOWITZ S, KOZAK C A, et al. NSCL-2: a basic domain helix-loop-helix gene expressed in early neurogenesis[J]. Cell Growth Differ, 1992, 3(3): 143–148. |
| [2] | HERBISON A E, PORTEOUS R, PAPE J R, et al. Gonadotropin-releasing hormone neuron requirements for puberty, ovulation, and fertility[J]. Endocrinology, 2008, 149(2): 597–604. DOI: 10.1210/en.2007-1139 |
| [3] | KRÜGER M, RUSCHKE K, BRAUN T. NSCL-1 and NSCL-2 synergistically determine the fate of GnRH-1 neurons and control necdin gene expression[J]. EMBO J, 2004, 23(21): 4353–4364. DOI: 10.1038/sj.emboj.7600431 |
| [4] | GOOD D J, PORTER F D, MAHON K A, et al. Hypogonadism and obesity in mice with a targeted deletion of the Nhlh2 gene[J]. Nat Genet, 1997, 15(4): 397–401. DOI: 10.1038/ng0497-397 |
| [5] | COGLIATI T, DELGADO-ROMERO P, NORWITZ E R, et al. Pubertal impairment in Nhlh2 null mice is associated with hypothalamic and pituitary deficiencies[J]. Mol Endocrinol, 2007, 21(12): 3013–3027. DOI: 10.1210/me.2005-0337 |
| [6] | JOHNSON S A, MARÍN-BIVENS C L, MIELE M, et al. The Nhlh2 transcription factor is required for female sexual behavior and reproductive longevity[J]. Horm Behav, 2004, 46(4): 420–427. DOI: 10.1016/j.yhbeh.2004.03.006 |
| [7] | NONNEMAN D, LENTS C, ROHRER G, et al. Genome-wide association with delayed puberty in swine[J]. Anim Genet, 2014, 45(1): 130–132. DOI: 10.1111/age.2013.45.issue-1 |
| [8] | TSUNODA T, TAKAGI T. Estimating transcription factor bindability on DNA[J]. Bioinformatics, 1999, 15(7-8): 622–630. |
| [9] | MATHELIER A, FORNES O, ARENILLAS D J, et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles[J]. Nucleic Acids Res, 2016, 44(D1): D110–D115. DOI: 10.1093/nar/gkv1176 |
| [10] |
高萍, 钟玉宜, 张爱玲. 母猪卵巢颗粒细胞的分离培养及鉴定[J]. 广东农业科学, 2014, 41(4): 131–135.
GAO P, ZHONG Y Y, ZHANG A L. Separation, culture and identification of sow ovarian granulose cells[J]. Guangdong Agricultural Sciences, 2014, 41(4): 131–135. (in Chinese) |
| [11] | LIU J Y, DU X, ZHOU J L, et al. MicroRNA-26b functions as a proapoptotic factor in porcine follicular Granulosa cells by targeting Sma-and Mad-related protein 4[J]. Biol Reprod, 2014, 91(6): 146. |
| [12] | GADE P, KALVAKOLANU D V. Chromatin immunoprecipitation assay as a tool for analyzing transcription factor activity[M]//VANCURA A. Transcriptional Regulation: Methods and Protocols. New York, NY: Springer, 2012: 85-104. |
| [13] | RODRÍGUEZ-UBREVA J, BALLESTAR E. Chromatin immunoprecipitation[M]//STOCKERT J C, ESPADA J, BLÁZQUEZ-CASTRO A. Functional Analysis of DNA and Chromatin. Totowa, NJ: Humana Press, 2014: 309-318. |
| [14] | MUELLER J K, KOCH I, LOMNICZI A, et al. Transcription of the human EAP1 gene is regulated by upstream components of a puberty-controlling Tumor Suppressor Gene network[J]. Mol Cell Endocrinol, 2012, 351(2): 184–198. DOI: 10.1016/j.mce.2011.12.004 |
| [15] | ROTH C L, MASTRONARDI C, LOMNICZI A, et al. Expression of a tumor-related gene network increases in the mammalian hypothalamus at the time of female puberty[J]. Endocrinology, 2007, 148(11): 5147–5161. DOI: 10.1210/en.2007-0634 |
| [16] | GRIFFITH G J, TRASK M C, HILLER J, et al. Yin-yang1 is required in the mammalian oocyte for follicle expansion[J]. Biol Reprod, 2011, 84(4): 654–663. DOI: 10.1095/biolreprod.110.087213 |
| [17] | MAALOUF S W, SMITH C L, PATE J L. Changes in MicroRNA expression during maturation of the bovine corpus luteum: regulation of luteal cell proliferation and function by microRNA-34a[J]. Biol Reprod, 2016, 94(3): 71. |
| [18] | SANDISON H E, USHER S, KARIMIANI E G, et al. PLK1 and YY1 interaction in follicular lymphoma is associated with unfavourable outcome[J]. J Clin Pathol, 2013, 66(9): 764–767. DOI: 10.1136/jclinpath-2013-201461 |
| [19] | MANDAL K, BADER S L, KUMAR P, et al. An integrated transcriptomics-guided genome-wide promoter analysis and next-generation proteomics approach to mine factor(s) regulating cellular differentiation[J]. DNA Res, 2017, 24(2): 143–157. |
| [20] | NALBANT D, WILLIAMS S C, STOCCO D M, et al. Luteinizing hormone-dependent gene regulation in Leydig cells may be mediated by CCAAT/enhancer-binding protein-β[J]. Endocrinology, 1998, 139(1): 272–279. DOI: 10.1210/endo.139.1.5663 |
| [21] | STERNECK E, TESSAROLLO L, JOHNSON P F. An essential role for C/EBPβ in female reproduction[J]. Genes Dev, 1997, 11(17): 2153–2162. DOI: 10.1101/gad.11.17.2153 |
| [22] | FAN H Y, LIU Z L, JOHNSON P F, et al. CCAAT/enhancer-binding proteins (C/EBP)-α and -β are essential for ovulation, luteinization, and the expression of key target genes[J]. Mol Endocrinol, 2011, 25(2): 253–268. DOI: 10.1210/me.2010-0318 |
| [23] | ZHEN Y H, WANG L, RIAZ H, et al. Knockdown of CEBPβ by RNAi in porcine granulosa cells resulted in S phase cell cycle arrest and decreased progesterone and estradiol synthesis[J]. J Steroid Biochem Mol Biol, 2014, 143: 90–98. DOI: 10.1016/j.jsbmb.2014.02.013 |
| [24] | CHRISTENSON L K, JOHNSON P F, MCALLISTER J M, et al. CCAAT/enhancer-binding proteins regulate expression of the human steroidogenic acute regulatory protein (StAR) gene[J]. J Biol Chem, 1999, 274(37): 26591–26598. DOI: 10.1074/jbc.274.37.26591 |
| [25] | PALL M, HELLBERG P, BRÄNNSTRÖM M, et al. The transcription factor C/EBP-β and its role in ovarian function; evidence for direct involvement in the ovulatory process[J]. EMBO J, 1997, 16(17): 5273–5279. DOI: 10.1093/emboj/16.17.5273 |
| [26] | REN Y A, LIU Z L, MULLANY L K, et al. Growth Arrest Specific-1 (GAS1) is a C/EBP target gene that functions in ovulation and corpus luteum formation in mice[J]. Biol Reprod, 2016, 94(2): 44. |
| [27] | DUPUIS L, SCHUERMANN Y, COHEN T, et al. Role of leptin receptors in granulosa cells during ovulation[J]. Reproduction, 2014, 147(2): 221–229. |



