2. 铜仁学院乌江学院, 铜仁 554300;
3. 山西农业大学动物科技学院, 太谷 030801
2. Wujiang College, Tongren University, Tongren 554300, China;
3. College of Animal Science and Veterinary Medicine, Shanxi Agricultural University, Taigu 030801, China
动物卵泡发育是一个复杂的过程,在卵泡发育的不同阶段,一些特定基因对卵泡的募集、选择和闭锁起到了关键调控作用[1]。S.M.Romereim等[2]利用基因芯片技术对牛卵巢DF中GCs和膜细胞(Theca cells, TCs)以及黄体中的大黄体细胞(Large luteal cells, LLCs)和小黄体细胞(Small luteal cells, SLCs)进行转录组分析,筛选出大量调控牛卵泡发育和黄体形成的基因;E.Terenina等[3]对猪健康和闭锁卵泡GCs进行表达谱分析,共获得1 684个重要调控基因,其中高表达基因287个,筛选出11个卵泡闭锁的标记基因。课题组前期也对PDF1和ODF1卵泡进行了Illumina平台测序,共获得42个上调基因和41个下调基因可能与牛卵泡发育直接相关[4]。牛卵泡发育过程中,卵泡发育出现偏差(Deviation)被认为是卵泡发育的重要生理阶段和转折点,其中ODF1卵泡将发育为DF,具有排卵的潜力,而PDF2卵泡可能发育为SF,并最终走向闭锁,本研究以牛卵巢PDF2和ODF1不同生理阶段卵泡作为样本筛选卵泡发育相关基因,随机选择差异表达基因qRT-PCR验证分析其在DF和SF中的表达谱,提高了卵泡发育基因筛选的准确性,为深入研究基因调控牛卵泡发育奠定了基础。
1 材料与方法 1.1 试验动物及样品采集选择9头正常发情的青年母牛,G6G技术(双排卵同期发情技术的改进方案,即注射PGF后, 18~24 h补加1次)同期发情,每天两次B超检测,并实时记录卵泡直径变化。分别在卵泡发育波出现偏差前和偏差后,3头牛分离ODF1卵泡,3头牛分离PDF2卵泡,另3头牛分离DF和SF。
1.2 试验方法 1.2.1 总RNA的抽提将分离的各卵泡分别置于预盛DPBS的平皿上,眼科剪一分为二后刮取GCs,移液枪转移到EP管中,2 000 r·min-1离心15 min,弃上清,并加入10倍体积RNAiso Plus,抽提总RNA。
1.2.2 转录组测序文库的构建转录组cDNA文库的构建方法详见课题组前期研究[4-5],Illumina HiSeq 2000平台进行混合测序。
1.2.3 差异表达基因筛选转录本差异表达基因的筛选参照S.Audic等[6]的研究方法,设定条件:RPKM值≥0.5,PDF2-RPKM与ODF1-RPKM双向比值≥2,经FDR校正(P < 0.05),获得差异表达基因。
1.2.4 差异表达基因GO分析应用DAVID 6.7在线分析软件(https://david.ncifcrf.gov/)输入差异表达基因列表,物种选择牛在线分析差异表达基因的功能聚类(P value < 0.05)并作图。
1.2.5 卵泡发育相关基因的筛选差异表达基因经GO功能富集分析后,结合Genecards(http://www.genecards.org/)基因功能在线查询系统筛选,并获得与牛卵泡发育直接相关的基因。
1.2.6 qRT-PCR验证分析从筛选出的卵泡发育相关基因中,随机选择5个基因,利用qRT-PCR进行转录组表达谱验证分析。qRT-PCR研究中,DF和SF颗粒细胞总RNA均为实验室前期获得,经反转录后,-20 ℃保存备用。引物设计参考NCBI已提交的牛(Bos taurus)相关基因序列,RPLP0作为内参基因,Primer 5.0软件设计,并送TaKaRa公司合成引物(表 1)。
|
|
表 1 荧光定量PCR引物序列 Table 1 Primers sequence of quantitative PCR |
各基因相对表达量采用ΔΔCT法计算,表达量数值经内参基因RPLP0校正,结果采用“均值±标准差(Mean±SD)”表示,SPSS 18.0统计软件进行显著性分析。
2 结果 2.1 PDF2和ODF1转录组差异表达基因的筛选Illumina平台测序结果经数据库比对,共获得35 325个基因;设定参数RPKM值≥0.5的表达基因共15 413个,其中表 2列出了15个高表达基因,由表 2可知,这些高表达基因中有参与转录调控的RN5-8S1,也有参与细胞发育和内分泌激素调控的基因,如INHA、INHBA和FST,这些基因在PDF2和ODF1中的表达量变化差异较小。
|
|
表 2 转录组PDF2和ODF1中15个表达量最高的基因 Table 2 Top 15 highly expressed genes in PDF2 and ODF1 follicles |
RPKM值≥0.5的表达基因经FDR(P < 0.05)校正后,设定PDF2-RPKM/ODF1-RPKM≥2,共获得383个下调的差异表达基因;设定ODF1-RPKM/PDF2-RPKM≥2,共获得268个上调的差异表达基因。其中差异表达倍数最高的20个基因及其功能注释见表 3。
|
|
表 3 转录组PDF2和ODF1中10个差异表达倍数最高的上调和下调基因及其功能 Table 3 Top 10 up-regulated and down-regulated genes with high differential expression fold in PDF2 and ODF1 and their functions |
利用DAVID 6.7在线分析软件对651个差异表达倍数(Fold change)≥2的基因进行GO富集性分析,共有484个基因与数据库中牛基因功能聚类数据相匹配,分为3大类111组:其中BP占41.66%,MF占17.24%,CC占41.10%(图 1)。经GO分析对基因归类注释后,便于筛选与牛卵泡发育相关的基因。如CHGA、SYTL4、NDUFAF2负调控胰岛素的分泌,而胰岛素对牛卵泡GCs增殖和雌激素分泌具有重要影响;参与细胞发育和周期调控的基因有45个,通过蛋白磷酸化参与信号转导的调控基因有19个(图 1A);调控跨膜受体蛋白激酶活性的基因有EPHA4、NTRK1、TGFBR3、KIT和EPHA2,CAV1、CTSH、FN1和PCOLCE2 4个基因参与肽酶催化活性的调节(图 1B);参与蛋白复合体组分的基因有92个,参与DNA包装复合体组分的基因有8个(图 1C)。
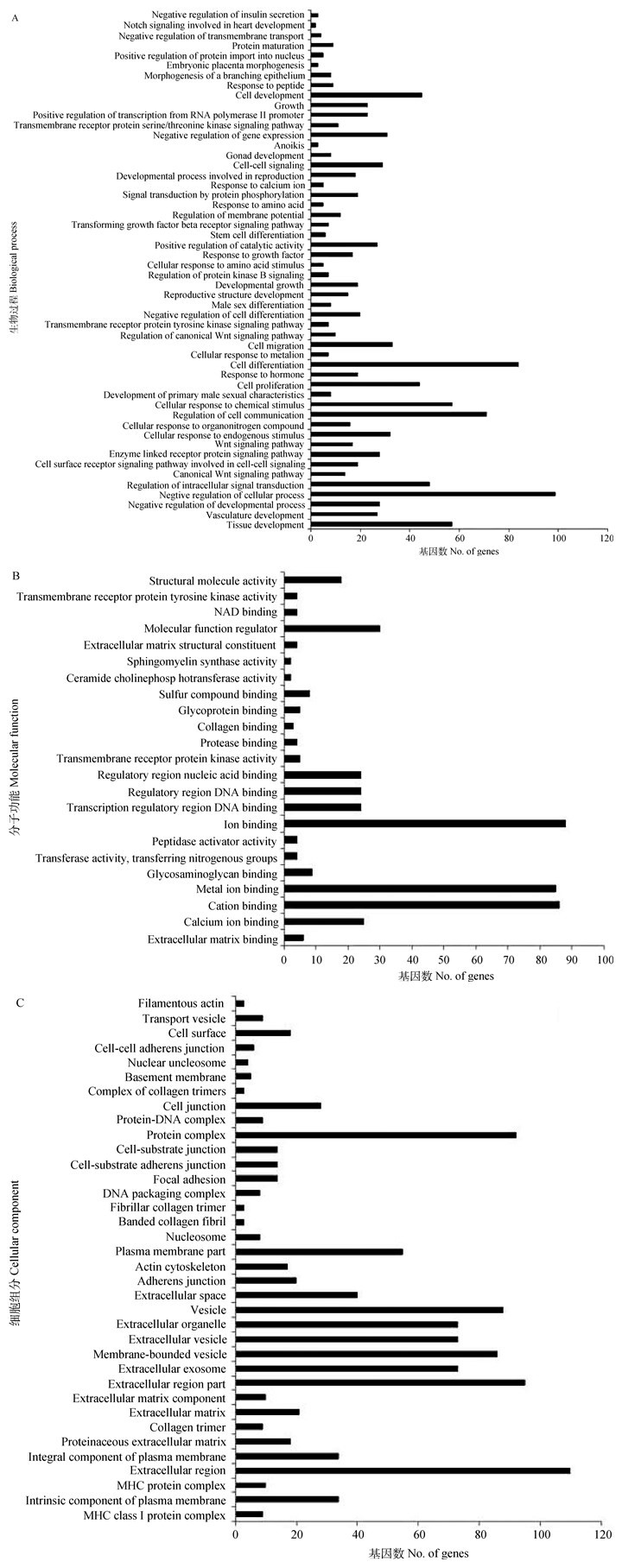
|
图 1 差异表达基因GO分析 Figure 1 GO analysis of differentially expressed genes |
经GO功能聚类分析后,结合Genecards人基因功能注释查询,共获得13个下调基因与牛卵泡发育直接相关,分别为MAPK13、GSTA5、SAFB2、WNT2B、EGR1、CHGA、KANK1、SESN3、PRICKLE1、ARID4B、TGFBR3、HBEGF和MAP2K6;获得17个与牛卵泡发育直接相关的上调基因,分别为CYP19A1、GREB1、SERPINE2、IGF2、NOTCH1、CYP17A1、SOCS3、LHCGR、SIX5、STK11、SYTL4、DACT1、CEBPB、COL3A1、NTRK1、DEPTOR和MYC,其表达差异倍数及功能注释见表 4。
|
|
表 4 牛卵泡发育相关基因 Table 4 The genes associated with bovine follicular development |
从表 4中随机选择MAPK13、CYP19A1、GREB1、SERPINE2和GSTA5共5个基因进行qRT-PCR表达量验证分析,由图 2可知,CYP19A1、GREB1和SERPINE2在DF中的表达量极显著高于SF(P < 0.01);MAPK13在SF中的表达量极显著高于DF(P < 0.01),GSTA5在SF中的表达量显著高于DF(P < 0.05)。qRT-PCR分析结果表明,各基因表达趋势与转录组测序结果相符,表明PDF2和ODF1转录组测序结果可信度较高。
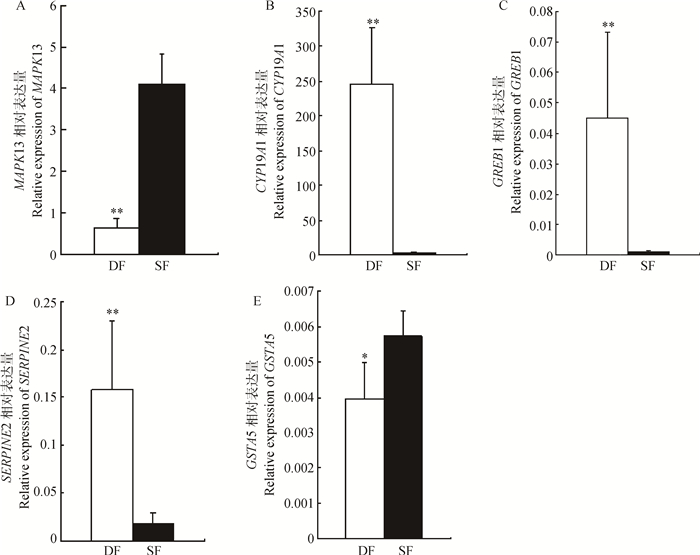
|
**. P<0.01;*.P<0.05.A. MAPK13; B.CYP19A1; C.GREB1;D.SERPINE2;E.GSTA5 图 2 qRT-PCR表达量验证分析 Figure 2 Real time PCR expression validation analysis |
高通量测序技术是转录组分析强有力的工具,在动物卵泡发育和成熟研究中已有大量报道[7-9]。通过全基因转录组分析,大量差异表达基因编码的信号分子如GDF9、BAX、BAD, NDUFA13、IFI6和CAV1与绵羊的多产性能密切相关[10];对牛卵泡发育过程中黄体生成素波峰前及波峰后的卵泡GCs转录组分析表明,调控细胞进程相关基因的高表达对母牛排卵具有重要意义[8],黄体生成素波峰后一组基因表达上调,对GCs黄体化发挥关键作用[11]。上述研究也表明,在细胞发育过程中转录组高通量测序是筛选关键调控基因的有效途径[12];同时,Illumina测序技术误差小,重复性也较好[13]。因此,本研究通过Illumina平台对牛卵泡发育波中PDF2和ODF1卵泡进行转录组测序,设定参数RPKM值≥0.5和表达差异倍数≥2,筛选差异表达基因,经GO富集性分析,FDR校正(P < 0.05)后,共获得13个下调基因和17个上调基因与牛卵泡发育直接相关,并通过qRT-PCR检测对转录组测序结果进行了验证分析。
MAPK13的相关研究较少,近年来,作为疾病特异性药物靶蛋白开始被人们关注,在体内表达具有严格的组织特异性,因此,对其功能的研究受到了限制[14-15]。这种受限制的表达模式表明该激酶可能以特定的生物学通路作为靶目标,如MAPK13调节胰岛素分泌和胰岛β细胞存活[16]。SAFB有SAFB1和SAFB2两种形式,调控细胞发育、增殖和凋亡[17]。研究表明,小鼠缺失SAFB1会导致IGF-1水平降低进而影响小鼠生长,通过降低雌二醇和黄体酮水平影响雌鼠生殖能力[18];反之,SAFB1过表达会使细胞周期S期变短,从而加速细胞凋亡[19]。EGR1是雌激素诱导的瞬时表达基因,以核磷蛋白或转录因子的形式调节细胞增殖或凋亡[20-21],同时EGR1调节信号对雌性生殖器官发育具有重要作用[22]。PRICKLE最初以平面细胞极性调控蛋白被识别[23],对小鼠的研究发现,PRICKLE1主要在发育胚胎的神经细胞中表达[24],基因敲除试验表明,PRICKLE1对外胚层和基底外侧极性的维持和建立具有重要作用[25]。ARID4B属于ARID家族,包含一个DNA连接活性区域螺旋-折叠-螺旋结构以及Tudor区域(TD)和chromo区域(CD)[26],TD和CD区域作为分子适配区域连接甲基化组蛋白,促进染色体重塑复合体的装配[27]。TGFB生长因子及其受体家族广泛参与细胞进程,包括增殖、迁移和分化,受体家族包括TGFBR1、TGFBR2和TGFBR3,对其功能研究多集中在心血管发育及其疾病调控方面[28]。
CYP19A1和CYP17A1是细胞色素P450超家族蛋白之一,参与类固醇激素的生物合成并在卵泡发育过程中发挥重要作用,研究表明,CYP19A1受到多重通路调控,包括GCs中雌激素受体以及FSH受体激活的cAMP/蛋白激酶A通路,这些通路可能决定了牛卵泡的优势化[29]。SERPINE2在牛卵泡发育波出现偏差前和偏差后卵泡GCs中均大量表达,提示SERPINE2对牛卵泡发育具有重要作用[4]。雌激素分泌和SERPINE2极显著相关,但雌激素处理体外培养的GCs不会改变SERPINE2的表达,FSH和生长因子可直接影响SERPINE2的分泌;SERPINE2属于抗凋亡因子,可能参与了牛卵泡闭锁的调控[30]。GREB1被证明是一种与染色质结合的雌激素受体共激活体,对雌激素受体介导的转录至关重要,因为它稳定了雌激素受体和其他辅助因子之间的相互作用;GREB1参与雌激素与其受体的结合,因此,GREB1也是一种潜在的临床生物标志物[31]。
4 结论经转录组差异表达基因筛选,共获得13个下调基因和17个上调基因与牛卵泡发育紧密相关,qRT-PCR表达量验证分析表明转录组数据可信度较高。
| [1] | BEG M A, BERGFELT D R, KOT K, et al. Follicle selection in cattle:dynamics of follicular fluid factors during development of follicle dominance[J]. Biol Reprod, 2002, 66(1): 120–126. DOI: 10.1095/biolreprod66.1.120 |
| [2] | ROMEREIM S M, SUMMERS A F, POHLMEIER W E, et al. Transcriptomes of bovine ovarian follicular and luteal cells[J]. Data Brief, 2017, 10: 335–339. DOI: 10.1016/j.dib.2016.11.093 |
| [3] | TERENINA E, FABRE S, BONNET A, et al. Differentially expressed genes and gene networks involved in pig ovarian follicular atresia[J]. Physiol Genomics, 2017, 49(2): 67–80. DOI: 10.1152/physiolgenomics.00069.2016 |
| [4] | LI P F, MENG J Z, LIU W Z, et al. Transcriptome analysis of bovine ovarian follicles at predeviation and onset of deviation stages of a follicular wave[J]. Int J Genomics, 2016, 2016: 347248. DOI: 10.1155/2016/3472748 |
| [5] |
李鹏飞, 孟金柱, 谢建山, 等. 牛卵泡ODF1与ODF2转录组发育相关基因筛选及表达差异分析[J]. 畜牧兽医学报, 2015, 46(11): 1961–1966.
LI P F, MENG J Z, XIE J S, et al. Screening and analyse study of genes associated with follicular development in bovine ODF1 and ODF2 transcript[J]. Acta Veterinaria et Zootechnica Sinica, 2015, 46(11): 1961–1966. (in Chinese) |
| [6] | AUDIC S, CLAVERIE J M. The significance of digital gene expression profiles[J]. Genome Res, 1997, 7(10): 986–995. DOI: 10.1101/gr.7.10.986 |
| [7] | XU Q, ZHAO W M, CHEN Y, et al. Transcriptome profiling of the goose (Anser cygnoides) ovaries identify laying and broodiness phenotypes[J]. PLoS One, 2013, 8(2): e55496. DOI: 10.1371/journal.pone.0055496 |
| [8] | HATZIRODOS N, IRVING-RODGERS H F, HUMMITZSCH K, et al. Transcriptome profiling of granulosa cells of bovine ovarian follicles during growth from small to large antral sizes[J]. BMC Genomics, 2014, 15: 24. DOI: 10.1186/1471-2164-15-24 |
| [9] | GILBERT I, ROBERT C, DIELEMAN S, et al. Transcriptional effect of the LH surge in bovine granulosa cells during the peri-ovulation period[J]. Reproduction, 2011, 141(2): 193–205. DOI: 10.1530/REP-10-0381 |
| [10] | MIAO X Y, LUO Q M, QIN X Y. Genome-wide transcriptome analysis of mRNAs and microRNAs in Dorset and Small Tail Han sheep to explore the regulation of fecundity[J]. Mol Cell Endocrinol, 2015, 402: 32–42. DOI: 10.1016/j.mce.2014.12.023 |
| [11] | GILBERT I, ROBERT C, VIGNEAULT C, et al. Impact of the LH surge on granulosa cell transcript levels as markers of oocyte developmental competence in cattle[J]. Reproduction, 2012, 143(6): 735–747. DOI: 10.1530/REP-11-0460 |
| [12] | LEI M M, CAI L P, LI H, et al. Transcriptome sequencing analysis of porcine granulosa cells treated with an anti-inhibin antibody[J]. Reprod Biol, 2017, 17(1): 79–88. DOI: 10.1016/j.repbio.2017.01.002 |
| [13] | CROUCHER N J, FOOKES M C, PERKINS T T, et al. A simple method for directional transcriptome sequencing using Illumina technology[J]. Nucleic Acids Res, 2009, 37(22): e148. DOI: 10.1093/nar/gkp811 |
| [14] | O'CALLAGHAN C, FANNING L J, BARRY O P. p38δ MAPK:emerging roles of a neglected isoform[J]. Int J Cell Biol, 2014, 2014: 272689. DOI: 10.1155/2014/272689 |
| [15] | RISCO A, CUENDA A. New insights into the p38γ and p38δ MAPK pathways[J]. J Signal Transduct, 2012, 2012: 520289. DOI: 10.1155/2012/520289 |
| [16] | SUMARA G, FORMENTINI I, COLLINS S, et al. Regulation of PKD by the MAPK p38δ in insulin secretion and glucose homeostasis[J]. Cell, 2009, 136(2): 235–248. DOI: 10.1016/j.cell.2008.11.018 |
| [17] | OESTERREICH S. Scaffold attachment factors SAFB1 and SAFB2:innocent bystanders or critical players in breast tumorigenesis?[J]. J Cell Biochem, 2003, 90(4): 653–661. DOI: 10.1002/(ISSN)1097-4644 |
| [18] | IVANOVA M, DOBRZYCKA K M, JIANG S, et al. Scaffold attachment factor B1 functions in development, growth, and reproduction[J]. Mol Cell Biol, 2005, 25(8): 2995–3006. DOI: 10.1128/MCB.25.8.2995-3006.2005 |
| [19] | TOWNSON S M, KANG K Y, LEE A V, et al. Structure-function analysis of the estrogen receptor α corepressor scaffold attachment factor-B1:identification of a potent transcriptional repression domain[J]. J Biol Chem, 2004, 279(25): 26074–26081. DOI: 10.1074/jbc.M313726200 |
| [20] | ADAMSON E D, MERCOLA D. Egr1 transcription factor:multiple roles in prostate tumor cell growth and survival[J]. Tumour Biol, 2002, 23(2): 93–102. DOI: 10.1159/000059711 |
| [21] | SUKHATME V P, CAO X M, CHANG L C, et al. A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, and after cellular depolarization[J]. Cell, 1988, 53(1): 37–43. DOI: 10.1016/0092-8674(88)90485-0 |
| [22] | RUSSELL D L, DOYLE K M H, GONZALES-ROBAYNA I, et al. Egr-1 induction in rat granulosa cells by follicle-stimulating hormone and luteinizing hormone:combinatorial regulation by transcription factors cyclic adenosine 3', 5'-monophosphate regulatory element binding protein, serum response factor, sp1, and early growth response factor-1[J]. Mol Endocrinol, 2003, 17(4): 520–533. DOI: 10.1210/me.2002-0066 |
| [23] | LIM B C, MATSUMOTO S, YAMAMOTO H, et al. Prickle1 promotes focal adhesion disassembly in cooperation with the CLASP-LL5β complex in migrating cells[J]. J Cell Sci, 2016, 129(16): 3115–3129. |
| [24] | OKUDA H, MIYATA S, MORI Y, et al. Mouse Prickle1 and Prickle2 are expressed in postmitotic neurons and promote neurite outgrowth[J]. FEBS Lett, 2007, 581(24): 4754–4760. DOI: 10.1016/j.febslet.2007.08.075 |
| [25] | TAO H, SUZUKI M, KIYONARI H, et al. Mouse prickle1, the homolog of a PCP gene, is essential for epiblast apical-basal polarity[J]. Proc Natl Acad Sci U S A, 2009, 106(34): 14426–14431. DOI: 10.1073/pnas.0901332106 |
| [26] | WILSKER D, PROBST L, WAIN H M, et al. Nomenclature of the ARID family of DNA-binding proteins[J]. Genomics, 2005, 86(2): 242–251. DOI: 10.1016/j.ygeno.2005.03.013 |
| [27] | NISHIBUCHI G, NAKAYAMA J I. Biochemical and structural properties of heterochromatin protein 1:understanding its role in chromatin assembly[J]. J Biochem, 2014, 156(1): 11–20. DOI: 10.1093/jb/mvu032 |
| [28] | DOETSCHMAN T, BARNETT J V, RUNYAN R B, et al. Transforming growth factor beta signaling in adult cardiovascular diseases and repair[J]. Cell Tissue Res, 2012, 347(1): 203–223. DOI: 10.1007/s00441-011-1241-3 |
| [29] | LUO W X, WILTBANK M C. Distinct regulation by steroids of messenger RNAs for FSHR and CYP19A1 in bovine granulosa cells[J]. Biol Reprod, 2006, 75(2): 217–225. DOI: 10.1095/biolreprod.105.047407 |
| [30] | CAO M J, NICOLA E, PORTELA V M, et al. Regulation of serine protease inhibitor-E2 and plasminogen activator expression and secretion by follicle stimulating hormone and growth factors in non-luteinizing bovine granulosa cells in vitro[J]. Matrix Biol, 2006, 25(6): 342–354. DOI: 10.1016/j.matbio.2006.05.005 |
| [31] | MOHAMMED H, D'SANTOS C, SERANDOUR A A, et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor[J]. Cell Rep, 2013, 3(2): 342–349. DOI: 10.1016/j.celrep.2013.01.010 |



