2. 中国兽医药品监察所, 北京 100086
2. China Institute of Veterinary Drugs Control, Beijing 100086, China
鸭源鸡杆菌(Gallibacterium anatis, G. anatis)属于巴氏杆菌科中的鸡杆菌属,该属包括数个种[1-2]。鸭源鸡杆菌包括溶血型和非溶血型两个生物型,然而,仅溶血型鸭源鸡杆菌在禽类中普遍流行,其感染往往造成蛋鸡输卵管炎和腹膜炎,产蛋率下降和死亡率升高[3-5]。随着近年来对该病原研究的深入,菌毛[6]、金属蛋白酶[7]、RTX毒素GtxA[8-9]、血凝素[10-11]、外膜囊泡[12]等一系列毒力因子得到鉴定和分析,但尚未见其外膜蛋白(outer membrane proteins,OMPs)生物学功能的相关研究报道。
OmpW是细菌外膜蛋白家族成员之一,属于小外膜蛋白家族,在革兰阴性菌中普遍存在,主要参与疏水性分子的跨膜转运[13-15]。研究显示OmpW有多种功能,如具有免疫原性[16]、参与调节细菌抗补体杀伤作用和抗吞噬作用[17-18]、增强细菌抗环境压力的能力[19-20]、与细菌毒力和致病性相关[21-22];另外,OmpW还与细菌耐药性有关[23],如沙门菌OmpW介导百草枯的外排[24];大肠杆菌OmpW可与多重耐药转运蛋白EmrE协同作用,介导抗菌药物外排,从而形成广泛耐药[25]。OmpW在鸭源鸡杆菌耐药性方面扮演什么角色,目前未见报道。本研究第一次利用自然转化法构建了鸭源鸡杆菌ompW缺失菌株(命名为ΔompW),分析了鸭源鸡杆菌亲本菌株及ΔompW的抗菌药物敏感性差异,以期了解OmpW在鸭源鸡杆菌耐药性中的作用。
1 材料与方法 1.1 菌株和质粒鸭源鸡杆菌PDS-RZ-1株(命名为RU)由本实验室分离、保存[26];质控菌ATCC27090和ATCC25922购自广东微生物菌种保藏中心;质粒pUC18、pMD18-T购自宝生物工程(大连)有限公司。
1.2 试剂和药品绵羊血琼脂平板购自郑州博赛生物工程有限公司;BHI培养基购自Difco公司;M-IV液体培养基组分均为分析纯级试剂,购自北京索莱宝科技有限公司;无维生素酪蛋白氨基酸购自美国BD公司;蛋白胨、酵母浸膏均购自北京奥博星生物技术有限责任公司;PCR扩增试剂及限制性内切酶购自宝生物工程(大连)有限公司;抗菌药物氯霉素、氟苯尼考、链霉素、庆大霉素、阿米卡星、环丙沙星、恩诺沙星、土霉素、四环素、红霉素、氨苄青霉素、喹乙醇均为国产原料药,符合中国药典或中国兽药典质量标准。
1.3 ompW基因缺失菌株的构建 1.3.1 引物参考GenBank中UMN179基因组中ompW基因及其上下游序列和质粒pBC KS+的序列信息,利用Primer 5.0设计5对引物(表 1)。引物由生工生物工程(上海)有限公司合成。
|
|
表 1 本研究所用引物 Table 1 PCR primers used in this study |
以鸭源鸡杆菌基因组为模板,分别以OmpW-U-F和OmpW-U-R,OmpW-D-F和OmpW-D-R为引物进行PCR扩增;以质粒pBC KS+为模板,CMP-F和CMP-R为引物进行PCR扩增。反应条件:95 ℃ 5 min;95 ℃ 30 s,56 ℃ 30 s,72 ℃ 60 s,30个循环;72 ℃ 10 min。PCR产物通过1%琼脂糖凝胶电泳检测,并送生工生物工程(上海)有限公司测序。纯化回收PCR产物,依次得到cmpr筛选标记片段和ompW上下游同源臂片段。
1.3.3 线性打靶DNA片段的制备分别用EcoR Ⅰ/BamH Ⅰ和BamH Ⅰ/Hind Ⅲ双酶切ompW上下游同源臂片段,用EcoR Ⅰ/BamH Ⅰ双酶切pUC18。然后将ompW上游同源臂和下游同源臂同时连接pUC18,得到重组质粒pUC18-U-D。将BamH Ⅰ酶切处理后的cmpr筛选标记片段连接经BamH Ⅰ单酶切的重组质粒pUC18-U-D,构建重组质粒pUC18-U-C-D。将重组质粒pUC18-U-C-D分别进行酶切鉴定、PCR及测序鉴定。以鉴定正确的重组质粒pUC18-U-C-D为模板,PSCX-F和PSCX-R为引物进行PCR扩增,反应条件:95 ℃ 5 min;95 ℃ 30 s,56 ℃ 30 s,72 ℃ 120 s,共30个循环,72 ℃ 10 min。PCR产物通过1%琼脂糖凝胶电泳检测,并送生工生物工程(上海)有限公司测序。纯化回收的PCR产物即为线性打靶DNA片段。
1.3.4 自然转化法缺失ompW基因参考G. Poje等[27]的方法制备鸭源鸡杆菌感受态细胞。1 mL感受态细胞中加入1 μg线性打靶DNA片段,轻轻混匀,37 ℃ 100 r·min-1孵育20 min。然后加2 mL BHI培养基,继续孵育90 min;4 ℃ 5 000 r·min-1离心4 min,弃上清, 用BHI培养基重悬菌体。在Cmpr(4 μg·mL-1)的BHI琼脂平板上37 ℃培养36 h,筛选出的阳性转化子,命名为ΔompW。
1.3.5 重组菌株的PCR鉴定对筛选的阳性转化子,以OmpW-QC-F和OmpW-QC-R为引物进行PCR检测,PCR产物通过1%琼脂糖凝胶电泳检测,并送生工生物工程(上海)有限公司测序。
1.3.6 外膜蛋白SDS-PAGE及Western blot分析将RU和ΔompW培养过夜作为种子液,按1:100转接到BHI液体培养基中,当培养至菌液OD600 nm=0.6时收集菌体。按S. W. Kim等[28]描述的超速离心法提取外膜蛋白。所得外膜蛋白进行SDS-PAGE及Western blot分析(自制的兔抗OmpW多克隆抗体作为一抗)。
1.4 菌株的生长形态及生长特性研究挑取鸭源鸡杆菌RU和ΔompW的单个菌落,分别接种于5 mL BHI液体培养基中,37 ℃ 200 r·min-1振荡过夜培养。第二天用接种环吊取菌液,分别接种于绵羊血琼脂平板,置于37 ℃恒温培养箱培养过夜,观察它们的菌落形态和大小。同时,第二天按1:100的稀释度接种于新鲜的BHI液体培养基中。分别在37 ℃ 200 r·min-1振荡培养,0 h(培养开始前)、每小时取样测OD600 nm值。做三次重复,取平均值。利用软件GraphPad Prism 6绘制细菌生长曲线。
1.5 MIC值测定采用微量肉汤稀释法[29]测定12种代表性抗菌药物对RU和ΔompW的MIC值。由于还没有针对鸭源鸡杆菌的操作标准和判定标准,因此本研究参照NCCLS(2013) 中巴氏杆菌的标准进行操作和结果判断。
2 结果 2.1 成功构建鸭源鸡杆菌ompW缺失菌株 2.1.1 获得cmpr筛选标记片段及ompW上下游同源臂片段分别以OmpW-U-F/OmpW-U-R和OmpW-D-F/OmpW-D-R为引物进行ompW基因上下游同源臂片段的PCR扩增,分别扩增出大小为1 000 bp左右的DNA片段;以CMP-F和CMP-R为引物进行cmpr筛选标记片段的PCR扩增,扩增出大小为1 200 bp左右的DNA片段,如图 1。同时测序结果表明扩增片段与预期相符。
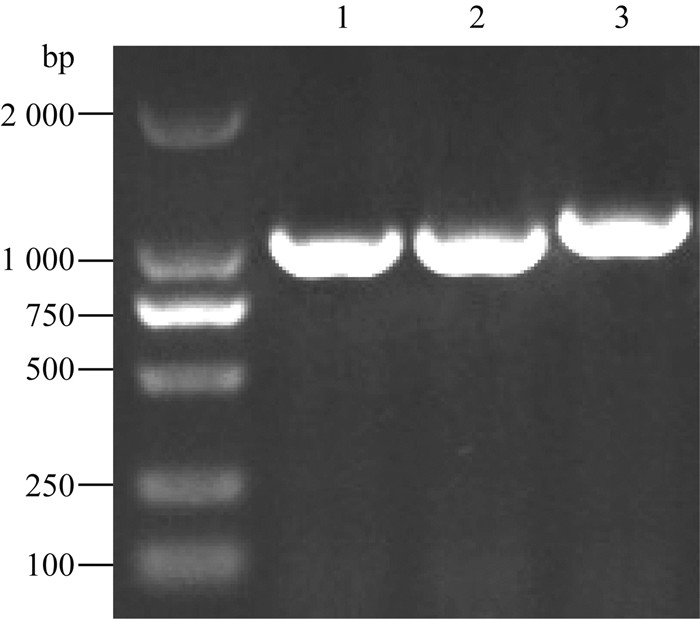
|
M.DL2000 DNA相对分子质量标准;1、2.分别是ompW上下游同源臂扩增产物;3. cmpr筛选标记片段扩增产物 M. DL2000 DNA marker; 1, 2. PCR product of upstream and downstream homologous arms of ompW gene, respectively; 3. PCR product of cmpr cassette 图 1 OmpW基因上下游同源臂片段及氯霉素抗性基因片段PCR扩增产物电泳 Figure 1 PCR amplification of upstream and downstream homologous arms of ompW and cmpr cassette |
分别用BamH Ⅰ单酶切,EcoR Ⅰ和Hind Ⅲ双酶切对重组质粒pUC18-U-C-D进行酶切鉴定,如图 2。同时重组质粒测序结果表明正确构建了重组质粒pUC18-U-C-D。
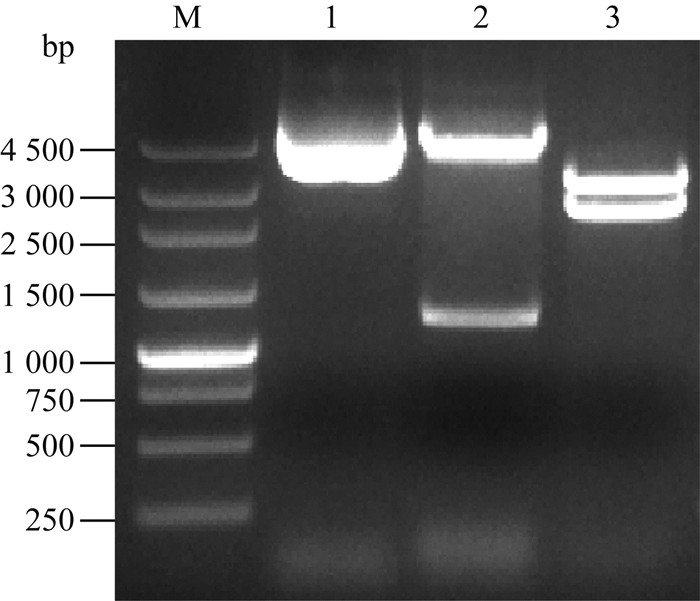
|
M. 250 bp DNA ladder;1.重组质粒pUC18-U-C-D;2. BamH Ⅰ酶切的重组质粒pUC18-U-C-D;3. EcoR Ⅰ和Hind Ⅲ双酶切的重组质粒pUC18-U-C-D M. 250 bp DNA ladder; 1. pUC18-U-C-D; 2. pUC18-U-C-D digested with BamH Ⅰ; 3. pUC18-U-C-D digested with EcoR Ⅰ and Hind Ⅲ 图 2 重组质粒pUC18-U-C-D的酶切鉴定 Figure 2 Analysis of recombinant plasmid by restriction enzymes |
以OmpW-QC-F/OmpW-QC-R为引物,分别以阳性转化子、亲本株基因组DNA为模板进行PCR检测,分别扩增出大小为1 300、700 bp左右DNA片段,大小与预期相符,如图 3。测序结果也证实筛选的阳性转化子为ompW缺失菌株ΔompW。
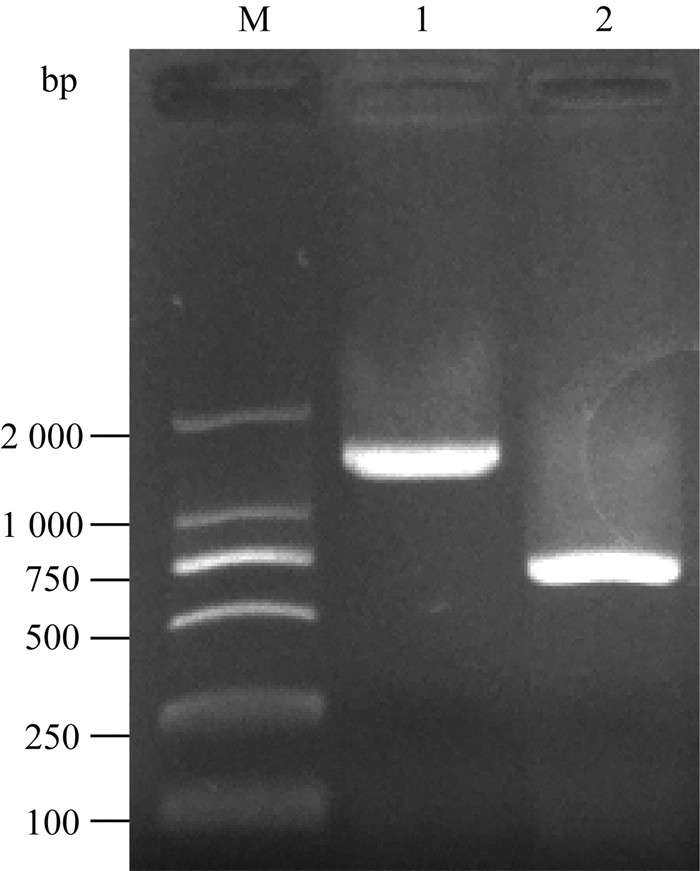
|
M. DL2000 DNA相对分子质量标准;1.阳性重组子;2.鸭源鸡杆菌亲本菌株 M. DL2000 DNA marker; 1. ΔompW; 2.G.anatis wild type 图 3 鸭源鸡杆菌ompW缺失菌株的PCR鉴定电泳图 Figure 3 PCR identification of G.anatis mutant ΔompW |
为进一步验证ompW是否成功敲除,利用超速离心提取RU和ΔompW外膜蛋白进行SDS-PAGE (图 4A),并利用抗OmpW多抗进行Western blot分析ΔompW是否不表达OmpW(图 4B)。结果显示ΔompW不表达该蛋白质,从而在蛋白质水平也证实成功构建了鸭源鸡杆菌ompW缺失菌株。
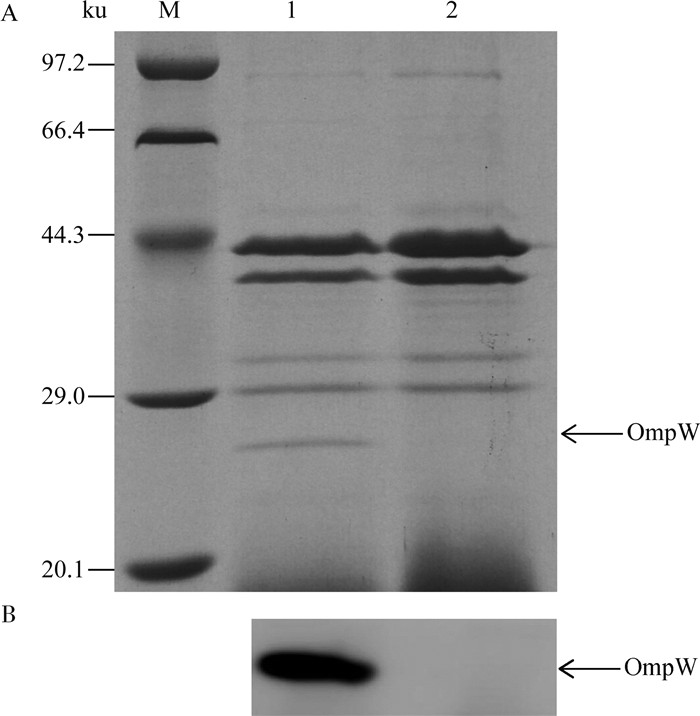
|
M.蛋白质低相对分子质量标准; 1.鸭源鸡杆菌亲本株菌RU;2.ΔompW M. Protein molecular weight marker (Low); 1. G. anatis wild type RU; 2. ΔompW 图 4 G.anatis RU和ΔompW外膜蛋白SDS-PAGE(A)及Western blot(B)分析 Figure 4 SDS-PAGE(A)and Western blot (B) analysis of membrane proteins extracted from G. anatis RU and ΔompW |
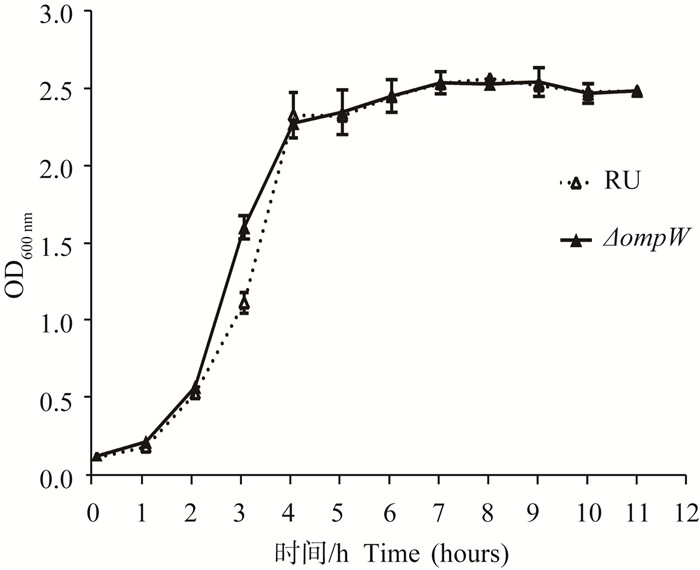
与RU相比,ΔompW菌落形态、大小没有明显肉眼可见差异(图 5),这表明ompW缺失并没有影响鸭源鸡杆菌的菌落形态及大小。每个小时测定一次菌液OD600 nm值,绘制细菌生长曲线;通过比较ΔompW与其亲本菌株RU的生长曲线,最初两者生长速度基本一致,进入对数生长期后,ΔompW的生长速度稍快;在进入平台期后,二者的菌量相当,总体生长趋势一致(图 6)。
|
图 5 鸭源鸡杆菌RU和ΔompW的生长形态 Figure 5 The growth morphology of G.anatis RU and ΔompW |
|
图 6 鸭源鸡杆菌RU与ΔompW的生长曲线 Figure 6 Culture curve of G.anatis RU and ΔompW |
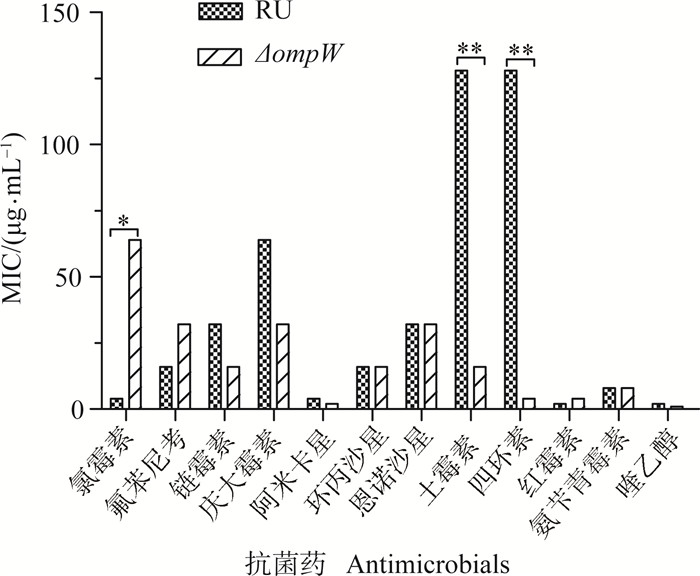
MIC值测定中,质控菌对参试抗菌药物均敏感,符合CLSI要求的标准。与亲本菌株相比,ompW缺失菌株ΔompW对土霉素和四环素的敏感性明显升高(图 7),它们的MIC值分别降低到1/8(16/128,P<0.01) 和1/32(4/128, P<0.01) 倍;而对氯霉素的敏感性明显降低,MIC值升高了16倍(64/4, P<0.05);其余参试抗菌药的MIC值无明显变化。
|
*.P < 0.05; **.P < 0.01 图 7 鸭源鸡杆菌RU和ΔompW对抗菌药的敏感性(MIC) Figure 7 The susceptibilities of RU and ΔompW to antibacterial agents |
自然感受态是一种使细菌具有从环境中摄入DNA并将其重组到细胞染色体的状态,在细菌中广泛存在[30-32]。细菌从营养丰富培养基到营养贫瘠培养基,或者从有氧环境到无氧环境的转变,诱导细菌一系列感受态相关元件表达,从而使细菌成为感受态,这些元件负责将环境中自由DNA黏附、转运及重组到细胞染色体[30, 33]。鸭源鸡杆菌作为巴氏杆菌科的一员,也具有转变为自然感受态的能力[34-35]。本研究利用MIV培养基诱导鸭源鸡杆菌呈现自然感受态,转化线性打靶DNA片段,经PCR、SDS-PAGE及Western blot分析,证实成功构建了鸭源鸡杆菌ompW缺失菌株。然而,由于鸭源鸡杆菌自身有一个或者多个质粒,可能由于这些质粒与常见表达质粒具有不相容性[1],或者其他一些未知原因,目前,未有鸭源鸡杆菌突变体回补菌株构建成功的报道,本研究同样也未获成功。
OmpW属于细菌外膜蛋白家族成员之一,在细菌多个方面扮演重要角色,其具有抗环境应激的能力[36-37]、免疫原性[16]、耐药性[25]且与细菌毒力相关[38]。鸭源鸡杆菌ompW缺失菌株构建成功不仅为下一步探讨鸭源鸡杆菌OmpW在细菌上述特性中的作用奠定研究基础,也为解析鸭源鸡杆菌其他蛋白质功能奠定了技术平台。
四环素类抗菌药通过干扰氨酰基-tRNA与细菌核糖体的结合来抑制细菌蛋白质合成,所以四环素必需穿过细胞膜才能与靶目标互作[39-40]。Mg2+-四环素复合物通过道南电位被吸附到细胞上,通过外膜孔蛋白孔通道或特殊的蛋白通道扩散穿过肠道革兰阴性细菌外膜,在外周胞质中积聚,复合物在外周胞质中解离,不带电荷的四环素成为弱亲脂性分子,可以扩散跨过细胞质膜脂质双分子层。细胞质中的四环素分子通常与镁离子结合成螯合物,进而与核糖体结合,影响蛋白质合成,发挥抑菌效应[39-41]。革兰阴性细菌主要利用外排作用获得四环素类药物抗性,转运蛋白能有效降低胞内四环素浓度。
OmpW是小孔蛋白家族成员,形成窄的疏水性通道,主要参与疏水性分子和离子的跨膜转运[13, 42]。本研究中,相比鸭源鸡杆菌亲本菌株,OmpW缺失菌株对土霉素和四环素(四环素类药物)的敏感性显著提高,二者的MIC值分别降低了8(128/16) 和32(128/4) 倍,而对其他参试抗菌药物(除氯霉素)的敏感性无明显变化,表明OmpW在鸭源鸡杆菌抗四环素类药物中发挥重要作用。2011年,A. M. Bojesen等[43]研究显示最常见的鸭源鸡杆菌四环素耐药因子为TetB,而TetB是四环素特异性外排泵蛋白[39]。2012年,吴贤斌等[44]发现敲除ompW基因后,大肠杆菌对硫酸新霉素和氨苄青霉素的敏感性大大增强,也有研究显示大肠杆菌的转运蛋白与外膜蛋白(Tolc,OmpW)协同作用发挥耐药活性[25, 45];F. Gil等[24]研究认为鼠伤寒杆菌OmpW可作为百草枯的外排通道。结合本研究结果,作者认为OmpW也可能是四环素类药物的一种外排通道,其丢失导致外排功能降低或丧失。
同时,细胞外膜孔蛋白数量、大小或选择性的变化影响抗菌药物的渗透能力[46],例如,孔蛋白丢失使很多细菌表现出明显的耐药性[47];淋病奈瑟氏菌孔蛋白编码基因的突变导致该菌对β-内酰胺类和四环素类药物产生抗性[48];也有研究指出外排泵蛋白与外膜蛋白协同作用将四环素排出,使细菌具有四环素抗性[49]。而本研究中鸭源鸡杆菌OmpW丢失没有提高ΔompW的耐药性,反而使其对土霉素和四环素的敏感性提高,这一结果间接提示鸭源鸡杆菌OmpW可能与外排蛋白协同发挥外排四环素类药物的作用。其中的具体作用机制还需要进一步研究确认。
4 结论首次用自然转化法构建了鸭源鸡杆菌ompW缺失菌株ΔompW;发现OmpW在鸭源鸡杆菌抗四环素类药物中发挥重要作用。
| [1] | CHRISTENSEN H, BISGAARD M, BOJESEN A M, et al. Genetic relationships among avian isolates classified as Pasteurella haemolytica, 'Actinobacillus salpingitidis' or Pasteurella anatis with proposal of Gallibacterium anatis gen. nov., comb. nov. and description of additional genomospecies within Gallibacterium gen. nov[J]. Int J Syst Evol Microbiol, 2003, 53(1): 275–287. DOI: 10.1099/ijs.0.02330-0 |
| [2] | BISGAARD M, KORCZAK B M, BUSSE H J, et al. Classification of the taxon 2 and taxon 3 complex of Bisgaard within Gallibacterium and description of Gallibacterium melopsittaci sp. nov., Gallibacterium trehalosifermentans sp. nov. and Gallibacterium salpingitidis sp. nov[J]. Int J Syst Evol Microbiol, 2009, 59(4): 735–744. DOI: 10.1099/ijs.0.005694-0 |
| [3] | PAUDEL S, LIEBHART D, HESS M, et al. Pathogenesis of Gallibacterium anatis in a natural infection model fulfils Koch's postulates: 1. Folliculitis and drop in egg production are the predominant effects in specific pathogen free layers[J]. Avian Pathol, 2014, 43(5): 443–449. DOI: 10.1080/03079457.2014.955782 |
| [4] |
王川庆, 陈陆, 杨霞, 等. 蛋鸡群卡氏杆菌感染情况的初步研究[J]. 河南农业科学, 2008(3): 97–100, 103.
WANG C Q, CHEN L, YANG X, et al. A premilinary study of Gallibacterium anantis infection in laying hens[J]. Journal of Henan Agricultural Sciences, 2008(3): 97–100, 103. (in Chinese) |
| [5] | SINGH S V, SINGH B R, SINHA D K, et al. Gallibacterium anatis: an emerging pathogen of poultry birds and domiciled birds[J]. J Vet Sci Technol, 2016, 7(3): 1000324. |
| [6] | BAGER R J, NESTA B, PORS S E, et al. The fimbrial protein FlfA from Gallibacterium anatis is a virulence factor and vaccine candidate[J]. Infect Immun, 2013, 81(6): 1964–1973. DOI: 10.1128/IAI.00059-13 |
| [7] | GARCÍA-GÓMEZ E, VACA S, PÉREZ-MÉNDEZ A, et al. Gallibacterium anatis-secreted metalloproteases degrade chicken IgG[J]. Avian Pathol, 2005, 34(5): 426–429. DOI: 10.1080/03079450500267866 |
| [8] | KRISTENSEN B M, FREES D, BOJESEN A M. Expression and secretion of the RTX-toxin GtxA among members of the genus Gallibacterium[J]. Vet Microbiol, 2011, 153(1-2): 116–123. DOI: 10.1016/j.vetmic.2011.05.019 |
| [9] |
李欢. 鸭源鸡杆菌流行株毒素蛋白GtxA表达及其部分生物学特性的研究[D]. 郑州: 河南农业大学, 2014.
LI H. The expression of protein GtxA and research of some biological characteristics of Gallibacterium anatis[D]. Zhengzhou: Henan Agricultural University, 2014. (in Chinese) http://cdmd.cnki.com.cn/Article/CDMD-10466-1014420759.htm |
| [10] | MONTES-GARCÍA J F, VACA S, VAZQUEZ-CRUZ C, et al. Identification of a hemagglutinin from Gallibacterium anatis[J]. Curr Microbiol, 2016, 72(4): 450–456. DOI: 10.1007/s00284-015-0969-5 |
| [11] | RAMIREZ-APOLINAR S, GUERRA-INFANTE F M, DE J DE HARO-CRUZ M, et al. Characterization of a Gallibacterium genomospecies 2 hemagglutinin[J]. J Anim Vet Adv, 2012, 11(4): 556–560. DOI: 10.3923/javaa.2012.556.560 |
| [12] | BAGER R J, PERSSON G, NESTA B, et al. Outer membrane vesicles reflect environmental cues in Gallibacterium anatis[J]. Vet Microbiol, 2013, 167(3-4): 565–572. DOI: 10.1016/j.vetmic.2013.09.005 |
| [13] | HONG H, PATEL D R, TAMM L K, et al. The outer membrane protein OmpW forms an eight-stranded β-barrel with a hydrophobic channel[J]. J Biol Chem, 2006, 281(11): 7568–7577. DOI: 10.1074/jbc.M512365200 |
| [14] | ALIZADEH J, RANJBAR R, KAMALI M, et al. Cloning of Vibrio cholerae outer membrane protein W in Pichia pastoris[J]. Iran J Microbiol, 2013, 5(3): 252–258. |
| [15] | HORST R, STANCZAK P, WÜTHRICH K. NMR polypeptide backbone conformation of the E. coli outer membrane protein W[J]. Structure, 2014, 22(8): 1204–1209. DOI: 10.1016/j.str.2014.05.016 |
| [16] | HUANG W W, WANG S J, YAO Y F, et al. OmpW is a potential target for eliciting protective immunity against Acinetobacter baumannii infections[J]. Vaccine, 2015, 33(36): 4479–4485. DOI: 10.1016/j.vaccine.2015.07.031 |
| [17] | WU X B, TIAN L H, ZOU H J, et al. Outer membrane protein OmpW of Escherichia coli is required for resistance to phagocytosis[J]. Res Microbiol, 2013, 164(8): 848–855. DOI: 10.1016/j.resmic.2013.06.008 |
| [18] | LI W Y, WEN L Y, LI C C, et al. Contribution of the outer membrane protein OmpW in Escherichia coli to complement resistance from binding to factor H[J]. Microb Pathog, 2016, 98: 57–62. DOI: 10.1016/j.micpath.2016.06.024 |
| [19] | XIAO M F, LAI Y, SUN J, et al. Transcriptional regulation of the outer membrane porin gene ompW reveals its physiological role during the transition from the aerobic to the anaerobic lifestyle of Escherichia coli[J]. Front Microbiol, 2016, 7: 799. |
| [20] | ASAKURA H, KAWAMOTO K, HAISHIMA Y, et al. Differential expression of the outer membrane protein W (OmpW) stress response in enterohemorrhagic Escherichia coli O157:H7 corresponds to the viable but non-culturable state[J]. Res Microbiol, 2008, 159(9-10): 709–717. DOI: 10.1016/j.resmic.2008.08.005 |
| [21] | XU D L, ZHANG J Y, LIU J, et al. Outer membrane protein OmpW is the receptor for typing phage VP5 in the Vibrio cholerae O1 El Tor biotype[J]. J Virol, 2014, 88(12): 7109–7111. DOI: 10.1128/JVI.03186-13 |
| [22] | WILKIE I W, HARPER M, BOYCE J, et al. Pasteurella multocida: diseases and pathogenesis[M]//AKTORIES K, ORTH J H C, ADLER B. Pasteurella Multocida. Berlin Heidelberg: Springer, 2012: 1-22. |
| [23] | LIN X M, YANG J N, PENG X X, et al. A novel negative regulation mechanism of bacterial outer membrane proteins in response to antibiotic resistance[J]. J Proteome Res, 2010, 9(11): 5952–5959. DOI: 10.1021/pr100740w |
| [24] | GIL F, IPINZA F, FUENTES J, et al. The ompW (porin) gene mediates methyl viologen (paraquat) efflux in Salmonella enterica serovar typhimurium[J]. Res Microbiol, 2007, 158(6): 529–536. DOI: 10.1016/j.resmic.2007.05.004 |
| [25] | BEKETSKAIA M S, BAY D C, TURNER R J. Outer membrane protein OmpW participates with small multidrug resistance protein member EmrE in quaternary cationic compound efflux[J]. J Bacteriol, 2014, 196(10): 1908–1914. DOI: 10.1128/JB.01483-14 |
| [26] |
郑鹿平. 我国鸡群鸡杆菌的首次分离鉴定及生物学特性初步研究[D]. 郑州: 河南农业大学, 2010.
ZHENG L P. The first isolation and identification of Gallibacterium anatis from fowls in China and preliminary research on its biological characterics[D]. Zhengzhou: Henan Agricultural University, 2010. (in Chinese) http://cdmd.cnki.com.cn/Article/CDMD-10466-2010270665.htm |
| [27] | POJE G, REDFIELD R J. Transformation of Haemophilus influenzae[M]//HERBERT M A, HOOD D W, MOXON E R. Haemophilus influenzae Protocols: Methods in Molecular Medicine. Totowa, NJ: Humana Press, 2003, 71: 57-70. |
| [28] | KIM S W, CHOI C H, MOON D C, et al. Serum resistance of Acinetobacter baumannii through the binding of factor H to outer membrane proteins[J]. FEMS Microbiol Lett, 2009, 301(2): 224–231. DOI: 10.1111/fml.2009.301.issue-2 |
| [29] | CLSI. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animal; approved standard-fourth edition, CLSI document VET01-A4[R]. Wayne, PA: Clinical and Laboratory Standards Institute, 2013. |
| [30] | JOHNSBORG O, ELDHOLM V, HÅVARSTEIN L S. Natural genetic transformation: prevalence, mechanisms and function[J]. Res Microbiol, 2007, 158(10): 767–778. DOI: 10.1016/j.resmic.2007.09.004 |
| [31] | LI J X, YUAN X F, XU L H, et al. Efficient construction of Haemophilus parasuis mutants based on natural transformation[J]. Can J Vet Res, 2016, 80(4): 281–286. |
| [32] | CHANG K C, YEH Y C, LIN T L, et al. Identification of genes associated with natural competence in Helicobacter pylori by transposon shuttle random mutagenesis[J]. Biochem Biophys Res Commun, 2001, 288(4): 961–968. DOI: 10.1006/bbrc.2001.5877 |
| [33] | SMITH H O, GWINN M L, SALZBERG S L. DNA uptake signal sequences in naturally transformable bacteria[J]. Res Microbiol, 1999, 150(9-10): 603–616. DOI: 10.1016/S0923-2508(99)00130-8 |
| [34] | KRISTENSEN B M, SINHA S, BOYCE J D, et al. Natural transformation of Gallibacterium anatis[J]. Appl Environ Microbiol, 2012, 78(14): 4914–4922. DOI: 10.1128/AEM.00412-12 |
| [35] | REDFIELD R J, FINDLAY W A, BOSSÉ J, et al. Evolution of competence and DNA uptake specificity in the Pasteurellaceae[J]. BMC Evol Biol, 2006, 6: 82. DOI: 10.1186/1471-2148-6-82 |
| [36] | BRAMBILLA L, MORÁN-BARRIO J, VIALE A M. Expression of the Escherichia coli ompW colicin S4 receptor gene is regulated by temperature and modulated by the H-NS and StpA nucleoid-associated proteins[J]. FEMS Microbiol Lett, 2014, 352(2): 238–244. DOI: 10.1111/fml.2014.352.issue-2 |
| [37] | MORALES E H, CALDERóN I L, COLLAO B, et al. Hypochlorous acid and hydrogen peroxide-induced negative regulation of Salmonella enterica serovar Typhimurium ompW by the response regulator ArcA[J]. BMC Microbiol, 2012, 12: 63. DOI: 10.1186/1471-2180-12-63 |
| [38] | PILSL H, SMAJS D, BRAUN V. Characterization of colicin S4 and its receptor, OmpW, a minor protein of the Escherichia coli outer membrane[J]. J Bacteriol, 1999, 181(11): 3578–3581. |
| [39] | GROSSMAN T H. Tetracycline antibiotics and resistance[J]. Cold Spring Harb Perspect Med, 2016, 6(4): a025387. DOI: 10.1101/cshperspect.a025387 |
| [40] | CHOPRA I, ROBERTS M. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance[J]. Microbiol Mol Biol Rev, 2001, 65(2): 232–260. DOI: 10.1128/MMBR.65.2.232-260.2001 |
| [41] | LI X Z, PLÉSIAT P, NIKAIDO H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria[J]. Clin Microbiol Rev, 2015, 28(2): 337–418. DOI: 10.1128/CMR.00117-14 |
| [42] | HATFALUDI T, AL-HASANI K, BOYCE J D, et al. Outer membrane proteins of Pasteurella multocida[J]. Vet Microbiol, 2010, 144(1-2): 1–17. DOI: 10.1016/j.vetmic.2010.01.027 |
| [43] | BOJESEN A M, VAZQUEZ M E, BAGER R J, et al. Antimicrobial susceptibility and tetracycline resistance determinant genotyping of Gallibacterium anatis[J]. Vet Microbiol, 2011, 148(1): 105–110. DOI: 10.1016/j.vetmic.2010.08.011 |
| [44] |
吴贤斌, 邹海杰, 田丽花, 等. 大肠杆菌外膜蛋白ompW基因敲除菌的构建及其对两种抗生素的敏感性[J]. 微生物学报, 2012, 52(8): 1021–1026.
WU X B, ZOU H J, TIAN L H, et al. Construction of ompW knock-out mutants of Escherichia coli to increase sensitivity to neomycinsulphate and ampicillin[J]. Acta Microbiologica Sinica, 2012, 52(8): 1021–1026. (in Chinese) |
| [45] | LOMOVSKAYA O, LEWIS K. Emr, an Escherichia coli locus for multidrug resistance[J]. Proc Natl Acad Sci U S A, 1992, 89(19): 8938–8942. DOI: 10.1073/pnas.89.19.8938 |
| [46] | YOSHIMURA F, NIKAIDO H. Diffusion of beta-lactam antibiotics through the porin channels of Escherichia coli K-12[J]. Antimicrob Agents Chemother, 1985, 27(1): 84–92. DOI: 10.1128/AAC.27.1.84 |
| [47] | WERNER V, SANDERS C C, SANDERS W E Jr, et al. Role of beta-lactamases and outer membrane proteins in multiple beta-lactam resistance of Enterobacter cloacae[J]. Antimicrob Agents Chemother, 1985, 27(4): 455–459. DOI: 10.1128/AAC.27.4.455 |
| [48] | GILL M J, SIMJEE S, AL-HATTAWI K, et al. Gonococcal resistance to β-lactams and tetracycline involves mutation in loop 3 of the porin encoded at the penB locus[J]. Antimicrob Agents Chemother, 1998, 42(11): 2799–2803. |
| [49] | SCHNAPPINGER D, HILLEN W. Tetracyclines: antibiotic action, uptake, and resistance mechanisms[J]. Arch Microbiol, 1996, 165(6): 359–369. DOI: 10.1007/s002030050339 |



