沙门菌是重要的人畜共患病病原之一,血清型众多。研究表明沙门菌对人和动物的致病性与血清型及其所携带的毒力基因有关[1],不同血清型的沙门菌感染谱有所差异[2-3],而不同动物、不同地区的优势血清型也不尽相同[4-5]。毒力基因编码的产物与细菌的致病力密切相关[6-7],沙门菌的毒力基因主要分布在毒力岛上,现目前已发现有23个毒力岛(SPI1~SPI23)存在于沙门菌上[8],其中前5个毒力岛存在于多种血清型菌株中,并含有与沙门菌的定植、存活、复制和侵袭等能力的相关毒力基因[9];此外,原噬菌体、毒力质粒上也存在与沙门菌致病力相关的毒力基因[10]。
目前关于鸡、猪和牛源沙门菌的病原流行病学调查报道很多[3, 11-12],但针对山羊源沙门菌的病原流行病学调查仅见澳大利亚L. Duffy等的报道[13],他们对分离自澳大利亚2个屠宰场121头山羊的179株沙门菌进行血清学鉴定,证实其分属8个血清型,其中圣保罗沙门菌、鼠伤寒沙门菌为优势血清型。近年来,我国山羊养殖业发展迅速,关于山羊沙门菌病的临床病例报道日渐增多[14-15],本研究对分离自四川省的山羊源沙门菌的血清型进行鉴定,并研究其部分毒力基因分布和其对小鼠的致病力,为山羊沙门菌病的防控提供参考。
1 材料与方法 1.1 菌株和实验动物42株待检沙门菌于2014—2016年分离自126份四川成都、简阳、北川、攀枝花、西昌5个地区的规模化羊场表观健康山羊的粪便;毒力基因质控菌株采用御城门沙门菌SWUN3736,携带本试验检测的15种毒力基因;均由西南民族大学动物医学实验室保存。
SPF级BALB/c雌性小鼠,体重为20 g±2 g,购自成都达硕实验动物有限公司。
1.2 培养基与试剂SS培养基和TSB培养基购自杭州微生物试剂有限公司,PCR反应试剂(10×Buffer、dNTP、rTaq酶)购自TaKaRa公司,质粒提取试剂盒[Plasimd Mini Kit 1(100)]和胶回收试剂盒[Gel Extraction Kit 1(200)]购自OMEGA公司。沙门菌分型血清购自泰国S & A公司。
1.3 沙门菌血清型鉴定42株山羊源沙门菌的血清型鉴定由四川省疾病预防控制中心完成,采用Kauffmann-White沙门菌抗原来判定血清型[16]。
1.4 沙门菌基因组DNA及质粒的提取将待检菌株和阳性对照菌株接种于SS培养基,37 ℃培养,收集24 h菌落采用酚-氯仿法提取基因组DNA,按照质粒提取试剂盒说明书提取质粒。
1.5 沙门菌毒力基因的检测 1.5.1 毒力基因及其引物本研究检测的15种毒力基因分别是位于毒力岛(SPI)上的11个基因(SPI-1 invA、sopE、orgA、avrA,SPI-2 ttrB、sseL,SPI-3 rhuM、mgtC,SPI-4 orfL,SPI-5, sopB、pipD)、原噬菌体基因sodC1、菌毛基因lpfC以及位于毒力质粒上的pefA、spvC基因,引物由擎科生物技术有限公司合成,引物信息见表 1。
|
|
表 1 沙门菌毒力基因的引物信息 Table 1 Primers for 15 virulence genes of Salmonella |
以御城门沙门菌作为阳性对照,以无模板对照作为阴性对照,采用相同的反应体系和条件,PCR扩增待检菌株的15种毒力基因:反应体系为25 μL:模板DNA(其中检测毒力质粒上pefA和spvC基因的模板为质粒DNA,检测其余13种毒力基因的模板为染色体DNA) 2 μL,上、下游引物各0.6 μL,10× PCR Buffer 2.5 μL,dNTP 2 μL,Taq DNA聚合酶0.2 μL,超纯水补足至25 μL;反应条件:95 ℃预变性5 min;95 ℃变性45 s,60 ℃退火45 s,72 ℃延伸45 s,共30个循环;72 ℃延伸10 min。PCR产物用2%琼脂糖凝胶电泳,凝胶成像。对阳性产物用胶回收试剂盒进行胶回收,并与pMD18-T载体连接,转化到DH5α感受态细胞,挑取克隆产物送擎科生物技术有限公司测序。
1.6 山羊源沙门菌对小鼠的致病性试验根据毒力基因检测结果,随机选取含有所检测的15种毒力基因的2株山羊源肠炎沙门菌(SWUN3810、SWUN3816)、2株布利丹沙门菌(SWUN3820、SWUN3842)、2株阿贡纳沙门菌(SWUN3802、SWUN3803)共6株菌对BALB/c小鼠致病性的初步研究,用TSB培养基进行增菌;对35只BALB/c小鼠随机分为7组,每组5只,用灌胃法对BALB/c小鼠进行攻毒试验,攻毒前12 h禁饮禁食,攻毒剂量为5×108 CFU·只-1,对照组灌胃相同剂量的培养基,攻毒后正常饲喂,观察记录小鼠的发病和死亡情况,并对发病死亡小鼠进行剖解和沙门菌的回收。
将山羊源肠炎沙门菌SWUN3816,接种于TSB培养基,37 ℃培养18 h,采用比浊法测定细菌浓度,采用平板计数法测定其菌落形成单位(CFU),用TSB培养基将菌液作10倍递增稀释,使其终浓度为102~108 CFU·mL-1。将35只小鼠随机分为7组,每组5只,于灌胃前12 h禁饮禁食,1~7组分别灌胃接种不同浓度的沙门菌,0.5 mL·只-1。灌胃后各组小鼠隔离饲养,自由采食和饮水,逐日观察并记录各组小鼠发病死亡情况,采用Bliss法计算LD50。
2 结果 2.1 山羊源沙门菌的血清型血清型鉴定结果显示,42株山羊源沙门菌分属4个血清型,分别为布利丹沙门菌23株、肠炎沙门菌15株、阿贡纳沙门菌3株和纽波特沙门菌1株,4种沙门菌所占比例分别为54.8%、35.7%、7.1%、2.4%,具体信息见表 2。
|
|
表 2 5个山羊场沙门菌分离率及其血清型 Table 2 Distribution of serotype of Salmonella strains from 5 goat farms |
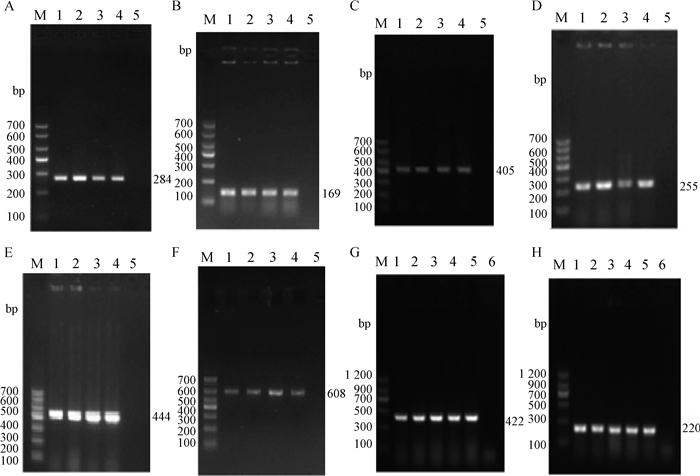
|
A~H分别为invA、sseL、pipD、orgA、sodC1、ttrB、avrA、sopB基因电泳(图A~F:M. Marker Ⅰ;1.阳性对照;2~4.部分样品检测结果;5.阴性对照;图G、H:M. Marker Ⅱ;1.阳性对照;2~5.部分样品检测结果;6.阴性对照) A-H show the electrophoresis of PCR products of invA, sseL, pipD, orgA, sodC1, ttrB, avrA, sopB genes (Fig.A-F:M. Marker Ⅰ; 1. Reference of SWUN3816; 2-4. Electrophoresis result of Salmonella strains; 5. Negative control; Fig.G, H: M. Marker Ⅱ; 1. Reference of SWUN3816; 2-5. Electrophoresis result of Salmonella strains; 6. Negative control) 图 1 部分基因电泳检测结果 Figure 1 The electrophoretogram of PCR products of partial genes |
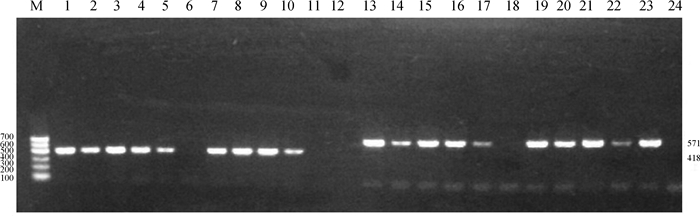
|
M. Marker Ⅰ;1. pefA基因阳性对照;2~11.部分样品pefA基因检测结果;12. pefA基因阴性对照;13. spvC基因阳性对照;14~23.部分样品spvC基因检测结果;24. spvC基因阴性对照 M. Marker Ⅰ; 1. pefA gene of SWUN3816 strain; 2-11. pefA genes of Salmonella strains; 12. Negative control of pefA gene; 13. spvC gene of SWUN3816 strain; 14-23. spvC genes of Salmonella strains; 24. Negative control of spvC gene 图 2 pefA 和spvC基因PCR产物电泳图 Figure 2 The electrophoretogram of the PCR product of pefA gene and spvC gene |
从图可见,从待检菌株中扩增出的毒力基因片段大小符合预期,测序结果证实为相应毒力基因的特异序列。
3株阿贡纳沙门菌中有两株(SWUN2802、SWUN3803)检测出pefA 和spvC基因。SWUN3802和SWUN3803的pefA 基因序列与肠炎沙门菌GQ258060.1中pefA基因序列相似度均为99%;而SWUN3802和SWUN3803的spvC基因序列与鼠伤寒沙门菌CP022071.1中spvC基因序列相似度分别为100%和99%。证明本试验所扩增基因确实为毒力质粒上的pefA 和spvC基因。
2.2.2 毒力基因的携带率PCR检测结果显示,42株待检菌株中,invA、orgA、pipD、sopB、lpfC、sopE、sseL、ttrB、rhuM、sodC1、avrA、mgtC、orfL基因的携带率分别为100%、100%、100%、100%、100%、97.6%、97.6%、88.1%、95.2%、92.9%、40.5%、40.5%、38.1%;毒力质粒上的标志性基因pefA和spvC存在于部分的阿贡纳沙门菌(2/3)、肠炎沙门菌(9/15)、布利丹沙门菌(9/23)中。
2.2.3 毒力基因的分布毒力基因的携带模式见表 3。
|
|
表 3 沙门菌毒力基因的携带模式 Table 3 Distribution mode of virulence genes in Salmonella |
从表 3可见,42株山羊源沙门菌毒力基因共有12种分布模式,其中以第7种模式即携带InvA-orgA-sopE-sseL-ttrB-rhuM-pipD-sopB-lpfC-sodC1这10种毒力基因的为主,占38%,其次是第1种模式携带15种毒力基因的分布模式,占33%;从血清型来看,布利丹沙门菌和肠炎沙门菌都有7种携带模式,均以第7种和第1种模式最多,分别占该血清型的47.8%、26.7%和26.1%、40%。3株阿贡那沙门菌有2种携带模式,第1种2株,第7种1株。
2.3 山羊源沙门菌对小鼠致病性试验情况对小鼠的致病性试验结果显示,携带15个毒力基因的2株阿贡纳沙门菌、2株布利丹沙门菌和2株肠炎沙门菌在5×108 CFU均能10 d内致死全部小鼠。攻毒12 h后小鼠开始出现扎堆、被毛杂乱、精神沉郁等现象,攻毒后30 h小鼠开始出现死亡。剖解发现病死小鼠肝表面有大量白色坏死灶、脾淤血、心出血、肺出血,小肠肠壁菲薄且充血等,且均能从心、肝、脾、肺这些实质器官中回收到沙门菌。
山羊源肠炎沙门菌SWUN3816在5×101和5×102 CFU剂量下对小鼠的致死率分别为20%和80%,在5×103~5×107 CFU剂量下对小鼠的致死率均为100%。采用Bliss方法计算得出该菌株对小鼠的LD50为1.6×102 CFU。
3 讨论本研究的42株山羊源沙门菌属于4个血清型,分别为布利丹沙门菌、肠炎沙门菌、阿贡纳沙门菌、纽波特沙门菌四种血清型;其中优势血清型为布利丹沙门菌和肠炎沙门菌,分别占54.8%和35.7%,二者是引起人食物中毒和沙门菌病的常见病原[24-25],所以山羊源沙门菌的公共卫生学意义值得关注。L. Duffy等[13]从澳大利亚2个屠宰场121头山羊上共分离到179株8个血清型,分别是圣保罗沙门菌、鼠伤寒沙门菌、切斯特沙门菌、阿贡纳沙门菌、阿德莱德沙门菌、病牛沙门菌、慕尼黑沙门菌及Anatum沙门菌,其中优势血清型为圣保罗沙门菌、鼠伤寒沙门菌。因此不同地域山羊沙门菌感染的血清型可能存在很大的差异。进一步加强不同区域山羊源沙门菌的调查,对丰富山羊源沙门菌的流行病学资料及对山羊沙门菌病的防控是有必要的。
沙门菌的毒力基因与其致病性密切相关。在本试验所检测的15个毒力基因中,invA、orgA、pipD、sopB、lpfC这5个毒力基因的携带率为100%,而sopE、sseL、ttrB、rhuM、sodC1这5个基因的携带率分别为97.6%、97.6%、88.1%、95.2%、92.9%。上述10种毒力基因在山羊源沙门菌的携带率很高,与人源、禽源和牦牛源沙门菌的结果相似[17, 23],说明这些基因普遍存在于不同宿主源的沙门菌中。其中,lpfC基因是菌毛基因,与细菌的黏附定植力相关[17];sodC1基因位于原噬菌体上,在细菌的胞内存活过程中起作用,与菌血症相关[22];而其余8种基因都位于毒力岛上,参与编码分泌系统相关蛋白,是重要的毒力因子[23]。细菌的致病性与其毒力基因的多少成正相关[7],本研究中山羊源沙门菌多数含有较多的毒力基因,这提示山羊源沙门菌可能存在较高的致病力。
而mgtC、avrA、orfL基因在本试验检测的菌株中携带率较低,分别为40.5%、40.5%和38.1%,显著低于人源、禽源和牦牛源沙门菌的携带率[17, 23, 26]。mgtC基因位于SPI3上,支持细菌耐受低Mg2+环境[27];orfL 位于SPI4上,促使细菌适应巨噬细胞内环境[28]。avrA基因存在于SPI1上,在细菌的致病过程中,可通过致弱肠上皮细胞等靶细胞,使得病原菌更加容易感染宿主[29]。这几个毒力基因在山羊源沙门菌与其他宿主源的沙门菌中的携带率有较大差别,是否是由于宿主不同有待进一步研究。
沙门菌的毒力质粒(virulence plasmid)在其全身性感染宿主过程中发挥重要作用,毒力质粒的携带与特定的血清型有关[1],pefA、spvC基因是沙门菌毒力质粒上特有的标志性基因[10, 17]。目前已报道的携带毒力质粒的沙门菌血清型10种,分别是鼠伤寒沙门菌、猪霍乱沙门菌、都柏林沙门菌、肠炎沙门菌、鸡沙门菌、S. Sendai、丙型副伤寒沙门菌、S. Abortusovis [30]以及布利丹沙门菌和御成门沙门菌[23]。本研究3株山羊源阿贡纳沙门菌中的2株携带毒力基因pefA、spvC,首次证实阿贡纳沙门菌携带毒力质粒[31-32],丰富了沙门菌基础生物学研究资料。
2012年M.C.Swearingen等[33]采用灌胃法感染未经抗生素处理的BALB/c小鼠, 测定了17种血清型32株沙门菌对小鼠的致病性,结果仅有2株沙门菌(1株鼠伤寒沙门菌和1株副伤寒沙门菌)可致死小鼠,其LD50分别为1.6×105和4.5×105 CFU;而另外30株菌均未致小鼠产生明显的临床症状。本试验中,随机抽取的含15种毒力基因的2株布利丹沙门菌、2株阿贡纳沙门菌和2株肠炎沙门菌在5×108 CFU剂量下均能全部致死未经抗生素处理的小鼠,显示出较强的毒力。特别值得注意的是,山羊源肠炎沙门菌SWUN3816经灌胃途径感染,可致死未经抗生素处理的BALB/c小鼠,其LD50为1.6×102 CFU,可见该菌株具有很强的毒力。肠炎沙门菌是本试验中沙门菌的优势血清型之一,而肠炎沙门菌、布利丹沙门菌和阿贡纳沙门菌都具有广泛的感染谱。因此,山羊源沙门菌的公共卫生学意义值得关注。
4 结论42株山羊源沙门菌鉴定出4种血清型,其中23株布利丹沙门菌、15株肠炎沙门菌、3株阿贡纳沙门菌、1株纽波特沙门菌;对受试菌株的15个毒力基因的检测结果表明有毒力基因携带模式共有12种,每株菌携带6~15种毒力基因;首次发现阿贡纳沙门菌携带毒力质粒。
| [1] | ANDINO A, HANNING I. Salmonella enterica: survival, colonization, and virulence differences among serovars[J]. Sci World J, 2015, 2015: 520179. |
| [2] | BUCKLE G C, WALKER C L F, BLACK R E. Typhoid fever and paratyphoid fever: systematic review to estimate global morbidity and mortality for 2010[J]. J Global Health, 2012, 2(1): 010401. |
| [3] |
邹明, 魏蕊蕊, 张纯萍, 等. 鸡源沙门氏菌的血清型、耐药性和耐药机制调查[J]. 农业生物技术学报, 2013, 21(7): 855–862.
ZOU M, WEI R R, ZHANG C P, et al. Investigation of serotype, antimicrobial susceptibility and resistant mechanisms of Salmonella isolated from Chickens (Gallus domesticus)[J]. Journal of Agricultural Biotechnology, 2013, 21(7): 855–862. (in Chinese) |
| [4] | PHILBEY A W, MATHER H A, GIBBONS J F, et al. Serovars, bacteriophage types and antimicrobial sensitivities associated with salmonellosis in dogs in the UK (1954-2012)[J]. Vet Rec, 2014, 174(4): 94. DOI: 10.1136/vr.101864 |
| [5] |
杨保伟, 张秀丽, 曲东, 等. 2007-2008陕西部分零售畜禽肉沙门氏菌血清型和基因型[J]. 微生物学报, 2010, 50(5): 654–660.
YANG B W, ZHANG X L, QU D, et al. Serotypic and genotypic characterization of Salmonella Serovars from retails meat in Shaanxi province (2007-2008)[J]. Acta Microbiologica Sinica, 2010, 50(5): 654–660. (in Chinese) |
| [6] |
王德宁. 鸡源沙门氏菌耐药性、致病性与毒力基因相关性分析[D]. 哈尔滨: 东北农业大学, 2014.
WANG D N. Correlation analysis among drug-resistance, pathogenicity and virulence genes of Salmonella isolated from chickens[D]. Harbin: Northeast Agricultural University, 2014. (in Chinese) http://cdmd.cnki.com.cn/Article/CDMD-10224-1014341363.htm |
| [7] | SUEZ J, PORWOLLIK S, DAGAN A, et al. Virulence gene profiling and pathogenicity characterization of non-typhoidal Salmonella accounted for invasive disease in humans[J]. PLoS One, 2013, 8(3): e58449. DOI: 10.1371/journal.pone.0058449 |
| [8] | HAYWARD M R, PETROVSKA L, JANSEN V A A, et al. Population structure and associated phenotypes of Salmonella enterica serovars Derby and Mbandaka overlap with host range[J]. BMC Microbiol, 2016, 16: 15. DOI: 10.1186/s12866-016-0628-4 |
| [9] | RYCHLIK I, KARASOVA D, SEBKOVA A, et al. Virulence potential of five major pathogenicity islands (SPI-1 to SPI-5) of Salmonella enterica serovar Enteritidis for chickens[J]. BMC Microbiol, 2009, 9: 268. DOI: 10.1186/1471-2180-9-268 |
| [10] | CASTILLA K S, FERREIRA C S A, MORENO A M, et al. Distribution of virulence genes sefC, pefA and spvC in Salmonella enteritidis phage type 4 strains isolated in Brazil[J]. Braz J Microbiol, 2006, 37(2): 135–139. DOI: 10.1590/S1517-83822006000200007 |
| [11] |
冯桂萍, 孙斌. 猪沙门氏菌病的流行、诊断与防治措施[J]. 现代畜牧科技, 2016(12): 75–76.
FENG G P, SUN B. Epidemics, diagnosis and prevention of Salmonella in pigs[J]. Technical Advisor for Animal Husbandry, 2016(12): 75–76. (in Chinese) |
| [12] |
王洋. 牛沙门氏菌病的流行特点[J]. 现代畜牧科技, 2015(10): 98.
WANG Y. Epidemiological characteristics of Salmonella in cattle[J]. Technical Advisor for Animal Husbandry, 2015(10): 98. DOI: 10.3969/j.issn.1673-1921.2015.10.079 (in Chinese) |
| [13] | DUFFY L, BARLOW R, FEGAN N, et al. Prevalence and serotypes of Salmonella associated with goats at two Australian abattoirs[J]. Lett Appl Microbiol, 2009, 48(2): 193–197. DOI: 10.1111/lam.2009.48.issue-2 |
| [14] |
吴良涛, 李涛, 张霞, 等. 贵州山羊流产的病原学调查[J]. 贵州农业科学, 2015, 43(8): 165–167.
WU L T, LI T, ZHANG X, et al. Investigation on etiology of goat abortion in Guizhou[J]. Guizhou Agricultural Sciences, 2015, 43(8): 165–167. (in Chinese) |
| [15] |
张治堂. 辽西白山羊沙门氏菌病的防治[J]. 特种经济动植物, 2017, 20(2): 19.
ZHANG Z T. Prevention and treatment of Salmonella in white goats of western Liaoning[J]. Special Economic Animal and Plant, 2017, 20(2): 19. (in Chinese) |
| [16] |
叶海梅, 刘红, 齐文岚, 等. 食物中毒沙门菌血清型、耐药性及毒力基因研究[J]. 热带医学杂志, 2017, 17(1): 11–14, 28.
YE H M, LIU H, QI W L, et al. Detection of serotype, drug resistance and virulence genes of food poisoning Salmonella[J]. Journal of Tropical Medicine, 2017, 17(1): 11–14, 28. (in Chinese) |
| [17] | ELEMFAREJI O I, THONG K L. Comparative virulotyping of Salmonellatyphi and Salmonella enteritidis[J]. Indian J Microbiol, 2013, 53(4): 410–417. DOI: 10.1007/s12088-013-0407-y |
| [18] | BORGES K A, FURIAN T Q, BORSOI A, et al. Detection of virulence-associated genes in Salmonella Enteritidis isolates from chicken in South of Brazil[J]. Pesq Vet Bras, 2013, 33(12): 1416–1422. DOI: 10.1590/S0100-736X2013001200004 |
| [19] | MEZAL E H, SABOL A, KHAN M A, et al. Isolation and molecular characterization of Salmonella enterica serovar Enteritidis from poultry house and clinical samples during 2010[J]. Food Microbiol, 2014, 38: 67–74. DOI: 10.1016/j.fm.2013.08.003 |
| [20] | GAL-MOR O, ELHADAD D, DENG W Y, et al. The Salmonella enterica PhoP directly activates the horizontally acquired SPI-2 Gene sseL and Is functionally different from a S. bongori Ortholog[J]. PLoS One, 2011, 6(5): e20024. DOI: 10.1371/journal.pone.0020024 |
| [21] | DOMINIK M M, GABRIELE M, REGINA K, et al. Next generation sequencing analysis of nine Corynebacterium ulcerans isolates reveals zoonotic transmission and a novel putative diphtheria toxin-encoding pathogenicity island[J]. Genome Med, 2014, 6(11): 113. DOI: 10.1186/s13073-014-0113-3 |
| [22] | AMMENDOLA S, AJELLO M, PASQUALI P, et al. Differential contribution of sodC1 and sodC2 to intracellular survival and pathogenicity of Salmonella enterica serovar Choleraesuis[J]. Microbes Infect, 2005, 7(4): 698–707. DOI: 10.1016/j.micinf.2005.01.005 |
| [23] |
张斌, 朱晓霞, 岳华, 等. 青藏高原部分地区牦牛源沙门菌血清型及毒力基因的调查[J]. 畜牧兽医学报, 2013, 44(7): 1167–1172.
ZHANG B, ZHU X X, YUE H, et al. The survey study on the serovar and virulent gene of yak Salmonella from some areas of Qinghai-Tibetan Plateau[J]. Acta Veterinaria et Zootechnica Sinica, 2013, 44(7): 1167–1172. (in Chinese) |
| [24] |
彭立昌, 马骏. 一起肠炎沙门菌引起的食物中毒调查分析[J]. 中国食品卫生杂志, 2017, 29(2): 233–237.
PENG L C, MA J. Investigation and analysis of an intestinal Salmonella enterica food poisoning incident[J]. Chinese Journal of Food Hygiene, 2017, 29(2): 233–237. (in Chinese) |
| [25] |
吴南卫, 邓瑶, 莫丽娟. 一起布利丹沙门菌食物中毒实验室检测及药敏分析[J]. 现代预防医学, 2015, 42(17): 3216–3217, 3224.
WU N W, DENG Y, MO L J. Detection of laboratory and drug sensitivity analysis on Salmonella food-poisoning from S. blegdam[J]. Modern Preventive Medicine, 2015, 42(17): 3216–3217, 3224. (in Chinese) |
| [26] |
赵方. 江苏地区禽源沙门菌的临床检测及相关特性的分析[D]. 扬州: 扬州大学, 2016.
ZHAO F. Clinic surveilling and characteristics of Salmonella isolates from avian of Jiangsu Province[D]. Yangzhou: Yangzhou University, 2016. (in Chinese) http://cdmd.cnki.com.cn/Article/CDMD-11117-1016284428.htm |
| [27] | LEE J W, LEE E J. Regulation and function of the Salmonella MgtC virulence protein[J]. J Microbiol, 2015, 53(10): 667–672. DOI: 10.1007/s12275-015-5283-1 |
| [28] | SÁNCHEZ-JIMÉNEZ M M, CARDONA-CASTRO N M, CANU N, et al. Distribution of pathogenicity islands among Colombian isolates of Salmonella[J]. J Infect Dev Ctries, 2010, 4(9): 555–559. |
| [29] | LU R, WU S P, ZHANG Y G, et al. Salmonella protein AvrA activates the STAT3 signaling pathway in colon cancer[J]. Neoplasia, 2016, 18(5): 307–316. DOI: 10.1016/j.neo.2016.04.001 |
| [30] | FOLEY S L, LYNNE A M. Food animal-associated Salmonella challenges: pathogenicity and antimicrobial resistance[J]. J Anim Sci, 2008, 86(14): 173–187. |
| [31] | ZIYATE N, KARRAOUAN B, KADIRI A, et al. Prevalence and antimicrobial resistance of Salmonella isolates in Moroccan laying hens farms[J]. J Appl Poul Res, 2016, 25(4): 539–546. DOI: 10.3382/japr/pfw036 |
| [32] | MCCUSKER M P, HOKAMP K, BUCKLEY J F, et al. Complete genome sequence of Salmonella enterica serovar agona pulsed-field type SAGOXB.0066, cause of a 2008 Pan-European Outbreak[J]. Genome Announc, 2014, 2(1): e01219–13. |
| [33] | SWEARINGEN M C, PORWOLLIK S, DESAI P T, et al. Virulence of 32 Salmonella strains in mice[J]. PLoS One, 2012, 7(4): e36043. DOI: 10.1371/journal.pone.0036043 |



