抗生素在抑制和杀灭细菌及各种致病微生物中有着不可替代的作用,细菌对抗生素产生耐药,导致一些药物对病原微生物感染的疗效降低甚至无效[1]。近几年的耐药研究显示,细菌对药物的主动外排在多重耐药(multidrug resistance,MDR)方面发挥着重要作用,即抗菌药物进入菌内,会激活细菌内膜的外排蛋白或调节蛋白,将药物排出,使细菌细胞内的药物浓度降低,从而导致药物对细菌的作用减弱或失去药效。在细菌细胞中,能将药物排出至细胞外的外排蛋白又被称作“外排泵”。由外排泵诱导的细菌耐药主要分为固有耐药、获得性耐药和表型性耐药[2]。
外排泵存在于大多数病原微生物,诱发外排蛋白发挥活性的机制各异,最终通过外排蛋白结合底物将其排出。已发现的细菌外排泵相关蛋白质,包括组成蛋白和调节蛋白。根据药物与蛋白质结合的受体分析法原理[3-4],将具有特异性结合一类抗生素的外排蛋白用于药物的残留检测,建立检测的新方法。
笔者拟综述各类抗生素在大部分革兰阳性菌和革兰阴性菌主要的外排蛋白,以及相关调节蛋白的调控,并分析与抗生素特异性结合的外排泵蛋白,总结外排泵研究方面存在的问题和发展趋势。旨在阐述外排蛋白的作用机制以及调控机制,进一步拓展外排泵相关蛋白的应用。
1 各类抗生素的外排泵组成蛋白和调控蛋白 1.1 细菌外排泵的分类和组成与细菌多重耐药有关的外排泵主要分为以下五类:主要易化家族(major facilitator super family,MFS)、小多药耐药家族(small multidrug resistance,SMR)、多药与毒物外排家族(multidrug and toxic microbial extrusion,MATE)、耐药结节分化家族(resistance nodulation and cell division,RND)和ATP结合盒家族(ATP binding cassette,ABC)[5]。其中,ABC类外排泵利用ATP水解释放的能量排出膜内有毒化合物[6],其余四类均通过质子进入膜内形成浓度差所产生的势能提供质子驱动力(H)为能量,从而将药物排至膜外[7]。
五类外排蛋白的组成如图 1所示。RND家族由经典的三联复合蛋白组成,即内膜转运蛋白(inner-membrane efflux pump, IMF)、外膜蛋白(outer membrane protein, OMF)和膜融合蛋白(membrane fusion protein, MFP)。该类外排蛋白复合物还存在于革兰阴性菌中的部分MFS和ABC家族。除三联复合蛋白外,其余的外排蛋白均由质膜上的单一跨膜蛋白组成,位于革兰阴性菌的细胞质内膜或者革兰阳性菌的单层质膜上。
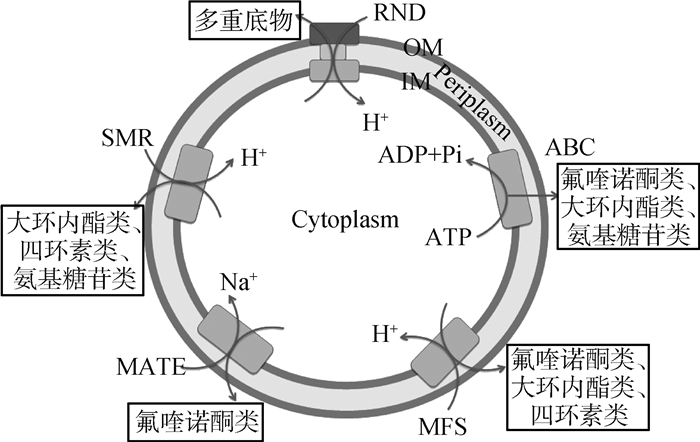
|
OM.外膜; IM.内膜 OM. Outer membrane; IM. Inner membrane 图 1 五类外排家族组成图 Figure 1 Schematic representation of the five types of bacterial efflux pumps |
外排泵的组成蛋白在发挥外排药物作用之前,均受到细菌胞内外排基因上游编码的调控蛋白所调控,从而诱发外排作用。调控蛋白种类繁多,根据其调控结果分为负调蛋白——阻遏蛋白(repressors)和正调蛋白——激活蛋白(activators),下面将详细介绍。
1.2 革兰阴性菌多底物外排泵及其调控革兰阴性菌外排泵几乎可以外排所有种类的抗生素(表 1)。大肠杆菌最重要的外排泵为AcrAB-TolC,属于RND家族。该外排泵由内膜蛋白AcrB、外膜通道蛋白TolC和膜连接蛋白AcrA组成。T. H. Schmidt等[8]的试验证明AcrB和TolC并不是蛋白质相互之间点对点进行对接,而是通过膜连接蛋白AcrA连接在一起,说明外排蛋白的组成缺一不可。该外排泵的调控蛋白有AcrR、MarA、Rob和SoxS[9],其中AcrR是局部调控,MarA、SoxS和Rob是全局调控。AcrR为负调因子[10],N端有DNA结合位点,C端有底物结合位点。菌体内没有药物存在时,AcrR与acrAB基因结合,抑制AcrAB蛋白表达。当药物进入菌体后,AcrR与药物结合引发构象改变,N端与DNA链分离,解除了acrAB基因的抑制作用,从而激活外排蛋白的表达(图 2)。MarA、SoxS和Rob[11]是转录调节蛋白,在氧化性应激等其他信号分子的作用下,结合在acrAB启动子附近的DNA结合位点,促进或抑制acrAB的转录,而从影响外排蛋白的合成。沙门菌也有AcrAB-TolC泵,RamA是沙门菌特有的调节因子,ramA不会受到外排底物的刺激而激活,而是当细菌受到外排泵抑制剂作用时诱导acrB失活,进而激活ramA,使RamA合成增加,促进AcrAB的表达[12]。
|
|
表 1 革兰阴性菌RND外排泵的组成 Table 1 The composition of RND efflux pumps present in Gram-negative organisms |
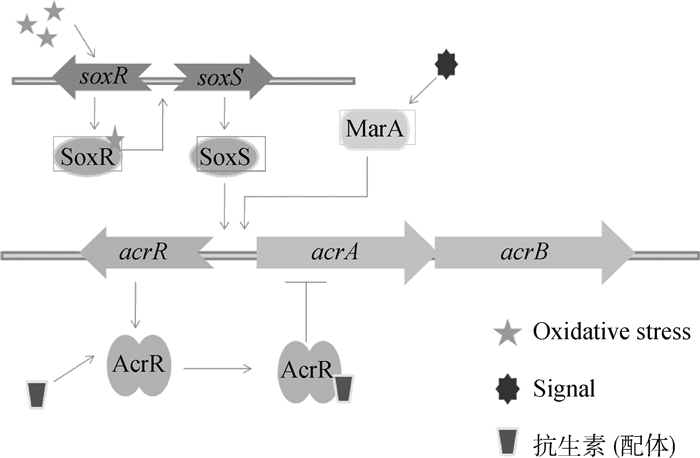
|
图 2 外排泵AcrAB-TolC的调控机制 Figure 2 AcrAB-TolC efflux pump regulation model |
鲍氏不动杆菌外排泵包括AdeABC、AdeFGH和AdeIJK,几乎外排所有抗生素。当AdeABC蛋白过度表达时,加强对四环素类药物的外排[13]。该类外排泵主要受到adeRS双组分调节系统的调节(图 3),该双组分调节系统由感应激酶蛋白AdeS和调节蛋白AdeR组成[14]。AdeS为组氨酸激酶感受器[15],在N端有2个跨膜结构域,接收外环境信号后,诱发自我磷酸化,同时将磷酸化信号转移到AdeR的天冬氨酸,从而激活AdeR。激活的AdeR结合于adeR和adeA之间的基因间隔区,诱导AdeABC泵的表达[16]。另外,AdeIJK外排泵受到BaeRS双组分调节系统的调节,特点与上述类似[17]。
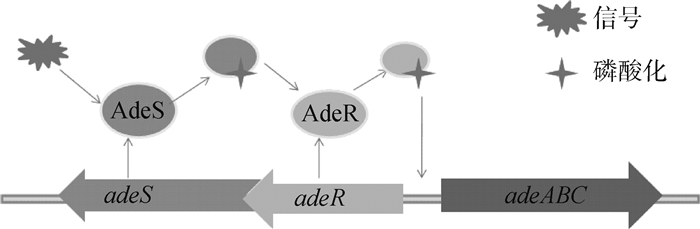
|
图 3 adeRS双组分调节系统的调控机制 Figure 3 adeRS, the two-component regulatory system and regulation model |
铜绿假单胞菌的外排泵主要是MexAB-OprM和MexXY-OprM[18]。当MexX和MexA蛋白过度表达时,加强该菌对氨基糖苷类和青霉素类的耐药[19]。该类外排泵的组成蛋白和AcrAB-TolC类似。在调节蛋白中,正调节蛋白有CpxR、BrlR、AmpR和MexT,负调节蛋白有MexR、NalD、MexT、RocA2和MexZ(图 4)。CpxR是全局调控蛋白,感受膜蛋白包括错误折叠的一系列细胞反应以及细菌外膜的压力,直接结合到mexA远端的启动子,激活MexAB-OprM的表达[20]。此外,CpxR和CpxA组成双组分调节系统,参与其他细菌的外排调节,如在大肠杆菌内CpxA作为细胞的压力蛋白,感受脂质双分子层的内外膜压力,将信号传递给CpxR,使其发生磷酸化[21]。BrlR[22]、AmpR[23]和MexT[24]的诱发机制尚未明确,但都具有DNA结合结构域,因此可以直接调节外排基因或其上游启动子区。负调蛋白MexR通过具有氧化还原活性的半胱氨酸之间的二硫键作用,而感受氧化性应激,激活MexR蛋白对mexAB基因的脱阻遏作用,使该基因表达[25]。RocA2是一种细菌菌毛的反应调节器,可以抑制MexAB-OprM泵的表达,说明该外排蛋白的合成和生物被膜的形成有一定的联系[26]。已有研究表明,MexZ蛋白的DNA结合位点发生突变,其底物结合位点与底物作用,促进MexXY蛋白的表达[27],NalD蛋白可以特异性结合新生霉素,通过MexAB-OprM将其排出[28]。
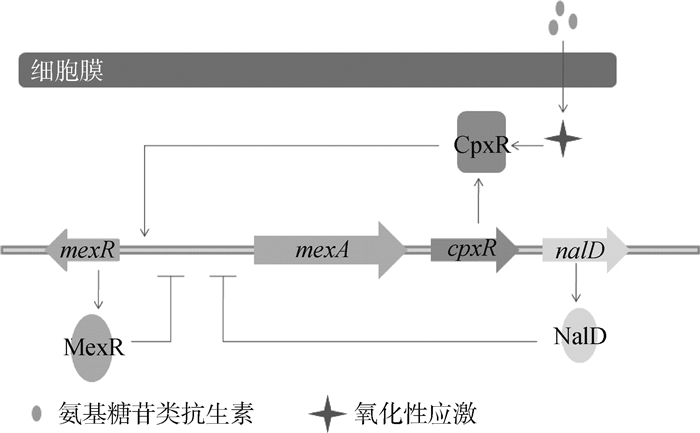
|
图 4 外排泵MexAB-OprM的调控机制 Figure 4 MexAB-OprM efflux pump regulation model |
恶臭假单胞菌的外排泵有TtgABC、TtgDEF和TtgGHI,受到转录调节蛋白TtgR的高度调节[29]。TtgR蛋白由DNA结合域以及底物结合域组成,TtgR结合ttgABC,可以抑制基因的转录。当药物结合于TtgR,引发其构象改变,解除对ttgABC的抑制,阻遏作用消失,TtgABC蛋白得以表达[30]。
嗜麦芽狭长平胞菌的外排泵有SmeABC、SmeDEF、SmeVWX、SmeIJK和SmeYZ,组成同大肠杆菌的AcrAB-TolC。外排底物不一,但都可以外排喹诺酮类抗菌药[31-32]。调节蛋白有SmeT和SmeRS,外排底物的差异主要是由于调节蛋白的不同造成的。SmeT是SmeDEF外排泵的阻遏调节蛋白,该调节蛋白末端的2个延伸片段组成DNA位点结合,同时位于活性中心的药物结合位点上的3个氨基酸主要是识别阳离子药物。SmeT结合药物后,其构象发生改变,对smeDEF的立体阻碍消失,使RNA聚合酶结合到DNA上,从而促进smeDEF表达[33-34]。SmeRS是双组分调节系统[35],smeRS位于smeA的上游,和鲍氏杆菌的AadRS的调节机制一致,组氨酸激酶感受蛋白SmeS和调节蛋白SmeR共同作用,调节SmeABC的表达。该菌还有多底物外排泵MacABCsm,属于ABC家族[36], 外排四环素类、氨基糖苷类和多黏菌素类抗生素,并受到MacR和MacS蛋白的双重调节。
肺炎克雷伯菌的RND外排泵主要是AcrAB和OqxAB,受到OqxR、RamR、RamA、RarA、SoxS和MarA蛋白的多重调节[37-39]。同大肠杆菌的调节类似,这些调节因子具有全局性,在上游信号的诱导下,直接或间接结合在外排基因上,抑制或促进外排蛋白的表达。RamA、RarA、SoxS和MarA四类是AraC类型的正调节蛋白[40],只有DNA结合结构域,激活oqxAB基因,促进OqxAB蛋白的表达,因四类调控蛋白不同的作用机制,使肺炎克雷伯菌对抗生素有着不同的敏感力(RamA>RarA>SoxS>MarA)[41]。此外,该类细菌还有KpnEF,是最新发现属于SMR家族的一种外排泵[42]。
淋病奈瑟球菌主要是MtrCDE外排泵,结构同大肠杆菌的AcrAB-TolC类似[43]。负调蛋白MtrR可以抑制MtrCDE的表达,编码该蛋白质的基因mtrR位于mtrC的上游,其功能和TetR家族蛋白类似[44]。MtrR既能结合于mtrR和mtrC之间的DNA序列上,抑制该外排泵基因的转录与翻译,也能结合在ponA-pilM的基因间隔区,ponA编码青霉素结合蛋白,pilM编码Ⅳ型菌毛分泌系统,形成青霉素通道蛋白,从而增加青霉素结合蛋白的细胞水平,加强对青霉素的耐药,说明MtrR在细菌内调节的全局性和非特异性[45]。另外,调节蛋白MtrA可以结合到MtrR结合位点上游的mtrC启动子区,促进MtrCDE泵的蛋白表达。这两种蛋白相反的调节结果,说明在一定条件的刺激下,其中一个蛋白与mtrCDE基因有更强的亲和性,进而调控该外排泵[46]。
1.3 革兰阴性菌单一底物外排泵及其调控除上述能同时外排多类抗生素的RND类外排泵外,MFS、ABC、MATE和SMR家族外排底物均较单一(表 2)。
|
|
表 2 革兰阴性菌非RND外排泵和革兰阳性菌外排泵的组成 Table 2 The composition of non-RND efflux pumps present in Gram-negative organisms and Gram-positive |
大肠杆菌TetA蛋白是内膜跨膜蛋白,由基因tetA编码,受到TetR蛋白的调控[47]。TetR是该外排泵的阻遏蛋白,四环素通过与Mg2+结合为阳离子复合物,从细菌外膜的膜孔蛋白进入到细胞质,结合到TetR阻遏蛋白操纵子复合物上,改变TetR的构象,使得TetR蛋白不能再结合到tetA的启动子区域,解除对tetA的抑制,使得TetA外排蛋白大量表达。四环素与Mg2+复合物作为该外排泵的底物,由质子驱动力提供能量,经TetA形成的跨膜蛋白排出(图 5)。我们将可以结合并诱导四环素类抗生素外排的调节蛋白统称为TetR转录调节阻遏蛋白家族,该家族包括经典的TetR,以及上述提到的AcrR、TtgR、MexZ、NalD和SmeT等[48]。除该类外排蛋白外,大肠杆菌的MFS家族还有EmrAB-TolC、MdfA和QepA[49-50],这些外排泵的组成蛋白都有药物的结合位点,如MdfA蛋白的氨基酸形成活性中心并结合氯霉素[51]。另外大肠杆菌ABC家族的MacAB-TolC泵,只外排大环内酯类抗生素,内膜蛋白MacB结合药物,ATP与其结合并提供转运能量。膜融合蛋白MacA内有脂多糖,在结合底物、刺激MacB上的ATP酶发挥活性有着至关重要的作用[52]。
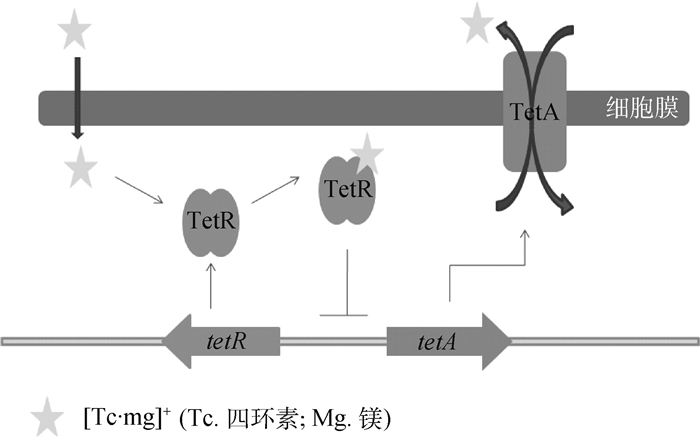
|
图 5 TetA家族的调控机制 Figure 5 TetA family regulation model |
奈瑟球菌具有与大肠杆菌一样的MacAB泵,专一外排大环内酯类抗生素[53]。FarAB-MtrE为经典的三联体复合蛋白,外排带有长链脂肪酸的抗生素,属于MFS家族,受到FarR的调节[54]。NorM是单一跨膜蛋白,属于MATE家族。当氟喹诺酮类抗菌药与细胞质内外排蛋白的氨基酸结合,加强离子(Na+)从胞外内流而进入NorM的阳离子结合区,使外排蛋白结构发现变化,结合的药物与氨基酸分离,并排出胞外[55-56]。
1.4 革兰阳性菌外排泵及其调控革兰阳性菌的外排泵有MFS、MATE、SMR和ABC四大类,主要是MFS类外排泵(表 2)。
金黄色葡萄球的MFS类外排泵,包括NorA、NorB、NorC和NorD(图 6),编码这几类外排蛋白的基因都位于染色体上,并表现出对氟喹诺酮类药物耐药[57]。该类外排蛋白由400个左右的氨基酸组成单一跨膜蛋白,受到MgrA和NorG,以及双组分调节蛋白ArlRS的调控。NorG是转录调节蛋白,MgrA和ArlRS是全局调控蛋白,均具有DNA结合结构域,可以结合到Nor等基因的启动子区,抑制或者促进蛋白表达[58-60]。另外,该菌还有MepA外排泵,调节蛋白MepR结合到mepA启动子的回文序列,抑制MepA的表达[61]。
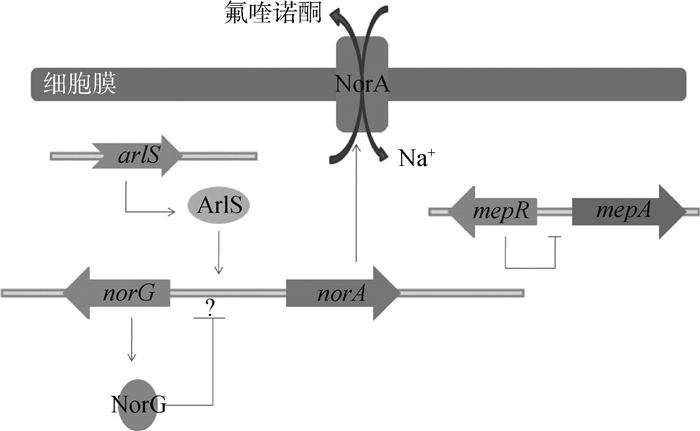
|
图 6 外排泵NorA的调控机制 Figure 6 NorA efflux pump regulation model |
肺炎链球菌的MFS类外排泵主要是Mef泵,以及ABC类的PatA和PatB泵。Mef外排泵包括Mef(A)和Mef(E)两类,由12个跨膜结构域组成(图 7)。编码该外排泵的基因位于染色体上,是革兰阳性菌专一外排大环内酯类抗生素的重要外排泵之一[62-63]。大环内酯类抗生素作用于核糖体上,诱导了在内酯环的第五位碳上多一个单糖,因此形成大环内酯类-核糖体-mRNA复合体,这个复合体的组成与mef的抗弱化转录调节一致[64]。因此,解除了对该外排泵基因的弱化,使得该外排泵蛋白得以表达,促进药物的外排[65]。PatA、PatB外排泵几乎可以外排所有的氟喹诺酮类抗菌药[66],其中对亲水性的氟喹诺酮类药物有更好的结合能力。patAB基因的转录终止是不依赖ρ因子的,patAB上游的3个单核苷酸突变会改变终止子结构,使patAB持续转录,因此促进该外排蛋白的翻译,增强药物的外排[67]。
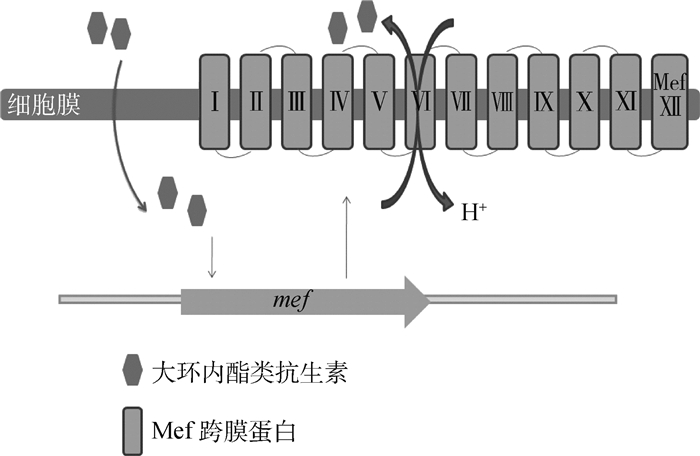
|
图 7 外排泵Mef的外排机制 Figure 7 Mef efflux pump excrete model |
单核细胞增生利斯特菌的MFS类外排泵主要有MdrL和Lde以及MATE类FepA外排泵。MdrL外排大环内酯类抗生素,受到LadR蛋白的负调控[68]。Lde外排四环素类和头孢类抗生素[69]。FepA单一外排氟喹诺酮类(如环丙沙星),FepR调节该外排蛋白的表达[70]。
除上述的致病微生物存在外排泵外,有益菌也有外排泵,如乳酸乳杆菌外排泵LmrA和LmrP,分别属于ABC和MFS家族,并表现出对大环内酯类、林可霉素类和链阳菌素的高度耐药[71]。
2 外排泵相关的抗生素结合蛋白在抗生素残留检测中的运用上述外排泵相关的抗生素结合蛋白,包括调节蛋白和组成蛋白,大多数与底物结合是非特异性的,它们可结合多种底物并促进外排,而少数能特异结合抗生素的调节蛋白,可被运用到抗生素的残留检测。
具体已经运用到抗生素残留检测的外排泵相关蛋白有TetR家族的TetR,TtgR调节蛋白。M. Espinosa-Urgel等[72]构建了基于恶臭假单胞菌的TtgABC外排泵调节蛋白TtgR的生物传感器,通过TtgR的活性中心与四环素、氯霉素和头孢类抗生素结合,通过各类抗生素的浓度不同,结合后形成的荧光强度不一,从而根据荧光强度测定不同抗生素含量。H. Hong等[73]利用大肠杆菌和不动杆菌,建立了基于TetA外排泵阻遏蛋白TetR的生物报告指示器,其活性中心可特异性结合多西环素。细菌的生物报告蛋白参与受启动子和报告基因调节的基因转录过程,被证明是一种可以检测多化合物的有力工具[74]。此前,L. H. Hansen等[75]构建TetR的绿色荧光蛋白,检测土壤中链霉菌产生的土霉素。
3 展望如今发现的外排抗生素的蛋白质很多,在众多的组成蛋白和调节蛋白中,能专一性结合抗生素并可以运用到残留检测中的蛋白质很少,大部分的调节蛋白具有全局性和非特异性。已经用于临床检测抗生素残留的蛋白质包括基于TtgR阻遏蛋白建立生物传感器,以及通过TetR蛋白建立生物报告指示器,检测四环素和氯霉素的残留。这两个阻遏蛋白都属于TetR家族,其蛋白质的结构特点是具有DNA结合结构域和底物结合结构域,有较为明确的药物结合位点,可以通过受体分析法用于抗生素的残留检测。除此类蛋白质家族外,其他外排泵相关蛋白质没有如此明显的结构特征,因此,还需要更多的试验研究外排泵蛋白质的相关性质,包括蛋白质的稳定性和活性,来判定是否能够通过上述方法用于残留检测,或者找到更合适的方法将其用于实践。
随着细菌耐药性逐年增加,发现细菌的外排机制愈发重要,需要更加清楚地研究引发细菌外排基因表达的机制,同时,将与抗生素专一性结合的外排泵相关蛋白质从众多的蛋白质中区分。因此,发现外排相关蛋白质与抗生素的结合位点,通过受体分析法,将其运用到药物的残留检测是以后研究的重点。
| [1] | HILTUNEN T, VIRTA M, LAINE A L. Antibiotic resistance in the wild:An eco-evolutionary perspective[J]. Philos Trans R Soc Lond B Biol Sci, 2017, 372(1712): 20160039. DOI: 10.1098/rstb.2016.0039 |
| [2] | HERNANDO-AMADO S, BLANCO P, ALCALDE-RICO M, et al. Multidrug efflux pumps as main players in intrinsic and acquired resistance to antimicrobials[J]. Drug Resist Updat, 2016, 28: 13–27. DOI: 10.1016/j.drup.2016.06.007 |
| [3] | LAMAR J, PETZ M. Development of a receptor-based microplate assay for the detection of beta-lactam antibiotics in different food matrices[J]. Anal Chim Acta, 2007, 586(1-2): 296–303. DOI: 10.1016/j.aca.2006.09.032 |
| [4] | AHMED S, NING J A, CHENG G Y, et al. Receptor-based screening assays for the detection of antibiotics residues-a review[J]. Talanta, 2017, 166: 176–186. DOI: 10.1016/j.talanta.2017.01.057 |
| [5] | BLANCO P, HERNANDO-AMADO S, REALES-CALDERON J A, et al. Bacterial multidrug efflux pumps:Much more than antibiotic resistance determinants[J]. Microorganisms, 2016, 4(1): 14. DOI: 10.3390/microorganisms4010014 |
| [6] | VERCHÈRE A, BROUTIN I, PICARD M. Photo-induced proton gradients for the in vitro investigation of bacterial efflux pumps[J]. Sci Rep, 2012, 2(1): 306. DOI: 10.1038/srep00306 |
| [7] | ADAM Y, TAYER N, ROTEM D, et al. The fast release of sticky protons:Kinetics of substrate binding and proton release in a multidrug transporter[J]. Proc Natl Acad Sci U S A, 2007, 104(46): 17989–17994. DOI: 10.1073/pnas.0704425104 |
| [8] | SCHMIDT T H, RAUNEST M, FISCHER N, et al. Computer simulations suggest direct and stable tip to tip interaction between the outer membrane channel TolC and the isolated docking domain of the multidrug RND efflux transporter AcrB[J]. Biochim Biophys Acta, 2016, 1858(7 Pt A): 1419–1426. |
| [9] | RUIZ C, LEVY S B. Regulation of acrAB expression by cellular metabolites in Escherichia coli[J]. J Antimicrob Chemother, 2014, 69(2): 390–399. DOI: 10.1093/jac/dkt352 |
| [10] | MANJASETTY B A, HALAVATY A S, LUAN C H, et al. Loop-to-helix transition in the structure of multidrug regulator AcrR at the entrance of the drug-binding cavity[J]. J Struct Biol, 2016, 194(1): 18–28. DOI: 10.1016/j.jsb.2016.01.008 |
| [11] | SATO T, YOKOTA S I, UCHIDA I, et al. Fluoroquinolone resistance mechanisms in an Escherichia coli isolate, HUE1, without quinolone resistance-determining region mutations[J]. Front Microbiol, 2013, 4: 125. |
| [12] | LAWLER A J, RICCI V, BUSBY S J W, et al. Genetic inactivation of acrAB or inhibition of efflux induces expression of ramA[J]. J Antimicrob Chemother, 2013, 68(7): 1551–1557. DOI: 10.1093/jac/dkt069 |
| [13] | YOON E J, COURVALIN P, GRILLOT-COURVALIN C. RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii:Major role for AdeABC overexpression and AdeRS mutations[J]. Antimicrob Agents Chemother, 2013, 57(7): 2989–2995. DOI: 10.1128/AAC.02556-12 |
| [14] | RICHMOND G E, EVANS L P, ANDERSON M J, et al. The Acinetobacter baumannii two-component system AdeRS regulates genes required for multidrug efflux, biofilm formation, and virulence in a strain-specific manner[J]. mBio, 2016, 7(2): e00430–16. |
| [15] | CAPRA E J, LAUB M T. Evolution of two-component signal transduction systems[J]. Annu Rev Microbiol, 2012, 66: 325–347. DOI: 10.1146/annurev-micro-092611-150039 |
| [16] | CHANG T Y, HUANG B J, SUN J R, et al. AdeR protein regulates adeABC expression by binding to a direct-repeat motif in the intercistronic spacer[J]. Microbiol Res, 2016, 183: 60–67. DOI: 10.1016/j.micres.2015.11.010 |
| [17] | LIN M F, LIN Y Y, LAN C Y. The role of the two-component system BaeSR in disposing chemicals through regulating transporter systems in Acinetobacter baumannii[J]. PLoS One, 2015, 10(7): e0132843. DOI: 10.1371/journal.pone.0132843 |
| [18] | TERZI H A, KULAH C, CIFTCIİ H. The effects of active efflux pumps on antibiotic resistance in Pseudomonas aeruginosa[J]. World J Microbiol Biotechnol, 2014, 30(10): 2681–2687. DOI: 10.1007/s11274-014-1692-2 |
| [19] | AGHAZADEH M, HOJABRI Z, MAHDIAN R, et al. Role of efflux pumps:MexAB-OprM and MexXY(-OprA), AmpC cephalosporinase and OprD porin in non-metallo-β-lactamase producing Pseudomonas aeruginosa isolated from cystic fibrosis and burn patients[J]. Infect Genet Evol, 2014, 24: 187–192. DOI: 10.1016/j.meegid.2014.03.018 |
| [20] | TIAN Z X, YI X X, CHO A, et al. CpxR Activates MexAB-OprM efflux pump expression and enhances antibiotic resistance in both laboratory and clinical nalB-type isolates of Pseudomonas aeruginosa[J]. PLoS Pathog, 2016, 12(10): e1005932. DOI: 10.1371/journal.ppat.1005932 |
| [21] | KELLER R, ARIÖZ C, HANSMEIER N, et al. The Escherichia coli envelope stress sensor CpxA responds to changes in lipid bilayer properties[J]. Biochemistry, 2015, 54(23): 3670–3676. DOI: 10.1021/acs.biochem.5b00242 |
| [22] | LIAO J L, SCHURR M J, SAUER K. The MerR-like regulator BrlR confers biofilm tolerance by activating multidrug efflux pumps in Pseudomonas aeruginosa biofilms[J]. J Bacteriol, 2013, 195(15): 3352–3363. DOI: 10.1128/JB.00318-13 |
| [23] | BALASUBRAMANIAN D, SCHNEPER L, MERIGHI M, et al. The regulatory repertoire of Pseudomonas aeruginosa AmpC β-lactamase regulator AmpR includes virulence genes[J]. PLoS One, 2012, 7(3): e34067. DOI: 10.1371/journal.pone.0034067 |
| [24] | MASEDA H, SAWADA I, SAITO K, et al. Enhancement of the mexAB-oprM efflux pump expression by a quorum-sensing autoinducer and its cancellation by a regulator, MexT, of the mexEF-oprN efflux pump operon in Pseudomonas aeruginosa[J]. Antimicrob Agents Chemother, 2004, 48(4): 1320–1328. DOI: 10.1128/AAC.48.4.1320-1328.2004 |
| [25] | CHEN H, YI C Q, ZHANG J, et al. Structural insight into the oxidation-sensing mechanism of the antibiotic resistance of regulator MexR[J]. EMBO Rep, 2010, 11(9): 685–690. DOI: 10.1038/embor.2010.96 |
| [26] | SIVANESON M, MIKKELSEN H, VENTRE I, et al. Two-component regulatory systems in Pseudomonas aeruginosa:An intricate network mediating fimbrial and efflux pump gene expression[J]. Mol Microbiol, 2011, 79(5): 1353–1366. DOI: 10.1111/mmi.2011.79.issue-5 |
| [27] | ALGUEL Y, LU D, QUADE N, et al. Crystal structure of MexZ, a key repressor responsible for antibiotic resistance in Pseudomonas aeruginosa[J]. J Struct Biol, 2010, 172(3): 305–310. DOI: 10.1016/j.jsb.2010.07.012 |
| [28] | CHEN W Z, WANG D, ZHOU W Q, et al. Novobiocin binding to NalD induces the expression of the MexAB-OprM pump in Pseudomonas aeruginosa[J]. Mol Microbiol, 2016, 100(5): 749–758. DOI: 10.1111/mmi.13346 |
| [29] | ALGUEL Y, MENG C X, TERÁN W, et al. Crystal structures of multidrug binding protein TtgR in complex with antibiotics and plant antimicrobials[J]. J Mol Biol, 2007, 369(3): 829–840. DOI: 10.1016/j.jmb.2007.03.062 |
| [30] | FERNANDEZ-ESCAMILLA A M, FERNANDEZ-BALLESTER G, MOREL B, et al. Molecular binding mechanism of TtgR repressor to antibiotics and antimicrobials[J]. PLoS One, 2015, 10(9): e0138469. DOI: 10.1371/journal.pone.0138469 |
| [31] | CHONG S Y, LEE K, CHUNG H S, et al. Levofloxacin efflux and smeD in clinical isolates of Stenotrophomonas maltophilia[J]. Microb Drug Resist, 2017, 23(2): 163–168. DOI: 10.1089/mdr.2015.0228 |
| [32] | SÁNCHEZ M B. Antibiotic resistance in the opportunistic pathogen Stenotrophomonas maltophilia[J]. Front Microbiol, 2015, 6: 658. |
| [33] | HERNÁNDEZ A, MATÉ M J, SÁNCHEZ-DÍAZ P C, et al. Structural and functional analysis of SmeT, the repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF[J]. J Biol Chem, 2009, 284(21): 14428–14438. DOI: 10.1074/jbc.M809221200 |
| [34] | HERNÁNDEZ A, RUIZ F M, ROMERO A, et al. The binding of triclosan to SmeT, the repressor of the multidrug efflux pump SmeDEF, induces antibiotic resistance in Stenotrophomonas maltophilia[J]. PLoS Pathog, 2011, 7(6): e1002103. DOI: 10.1371/journal.ppat.1002103 |
| [35] | WU C J, HUANG Y W, LIN Y T, et al. Inactivation of SmeSyRy two-component regulatory system inversely regulates the expression of SmeYZ and SmeDEF efflux pumps in Stenotrophomonas maltophilia[J]. PLoS One, 2016, 11(8): e0160943. DOI: 10.1371/journal.pone.0160943 |
| [36] | LIN Y T, HUANG Y W, LIOU R S, et al. MacABCsm, an ABC-type tripartite efflux pump of Stenotrophomonas maltophilia involved in drug resistance, oxidative and envelope stress tolerances and biofilm formation[J]. J Antimicrob Chemother, 2014, 69(12): 3221–3226. DOI: 10.1093/jac/dku317 |
| [37] | BIALEK-DAVENET S, LAVIGNE J P, GUYOT K, et al. Differential contribution of AcrAB and OqxAB efflux pumps to multidrug resistance and virulence in Klebsiella pneumoniae[J]. J Antimicrob Chemother, 2015, 70(1): 81–88. DOI: 10.1093/jac/dku340 |
| [38] | KÄLLMAN O, MOTAKEFI A, WRETLIND B, et al. Cefuroxime non-susceptibility in multidrug-resistant Klebsiella pneumoniae overexpressing ramA and acrA and expressing ompK35 at reduced levels[J]. J Antimicrob Chemother, 2008, 62(5): 986–990. DOI: 10.1093/jac/dkn296 |
| [39] | VELEBA M, HIGGINS P G, GONZALEZ G, et al. Characterization of RarA, a novel AraC family multidrug resistance regulator in Klebsiella pneumoniae[J]. Antimicrob Agents Chemother, 2012, 56(8): 4450–4458. DOI: 10.1128/AAC.00456-12 |
| [40] | SANTIAGO A E, YAN M B, TRAN M, et al. A large family of anti-activators accompanying XylS/AraC family regulatory proteins[J]. Mol Microbiol, 2016, 101(2): 314–332. DOI: 10.1111/mmi.2016.101.issue-2 |
| [41] | KAO C Y, CHEN C A, LIU Y F, et al. Molecular characterization of antimicrobial susceptibility of Salmonella isolates:First identification of a plasmid carrying qnrD or oqxAB in Taiwan[J]. J Microbiol Immunol Infect, 2017, 50(2): 214–223. DOI: 10.1016/j.jmii.2015.03.004 |
| [42] | SRINIVASAN V B, RAJAMOHAN G. KpnEF, a new member of the Klebsiella pneumoniae cell envelope stress response regulon, is an SMR-type efflux pump involved in broad-spectrum antimicrobial resistance[J]. Antimicrob Agents Chemother, 2013, 57(9): 4449–4462. DOI: 10.1128/AAC.02284-12 |
| [43] | GOLPARIAN D, SHAFER W M, OHNISHI M, et al. Importance of multidrug efflux pumps in the antimicrobial resistance property of clinical multidrug-resistant isolates of Neisseria gonorrhoeae[J]. Antimicrob Agents Chemother, 2014, 58(6): 3556–3559. DOI: 10.1128/AAC.00038-14 |
| [44] | JOHNSON P J T, SHAFER W M. The transcriptional repressor, MtrR, of the mtrCDE efflux pump operon of Neisseria gonorrhoeae can also serve as an activator of "off Target" gene (glnE) expression[J]. Antibiotics (Basel), 2015, 4(2): 188–197. DOI: 10.3390/antibiotics4020188 |
| [45] | FOLSTER J P, DHULIPALA V, NICHOLAS R A, et al. Differential regulation of ponA and pilMNOPQ expression by the MtrR transcriptional regulatory protein in Neisseria gonorrhoeae[J]. J Bacteriol, 2007, 189(13): 4569–4577. DOI: 10.1128/JB.00286-07 |
| [46] | ZALUCKI Y M, DHULIPALA V, SHAFER W M. Dueling regulatory properties of a transcriptional activator (MtrA) and repressor (MtrR) that control efflux pump gene expression in Neisseria gonorrhoeae[J]. mBio, 2012, 3(6): e00446–12. |
| [47] | MØLLER T S B, OVERGAARD M, NIELSEN S S, et al. Relation between tetR and tetA expression in tetracycline resistant Escherichia coli[J]. BMC Microbiol, 2016, 16: 39. DOI: 10.1186/s12866-016-0649-z |
| [48] | RAMOS J L, MARTÍNEZ-BUENO M, MOLINA-HENARES A J, et al. The TetR family of transcriptional repressors[J]. Microbiol Mol Biol Rev, 2005, 69(2): 326–356. DOI: 10.1128/MMBR.69.2.326-356.2005 |
| [49] | TANABE M, SZAKONYI G, BROWN K A, et al. The multidrug resistance efflux complex, EmrAB from Escherichia coli forms a dimer in vitro[J]. Biochem Biophys Res Commun, 2009, 380(2): 338–342. DOI: 10.1016/j.bbrc.2009.01.081 |
| [50] | YAMANE K, WACHINO J I, SUZUKI S, et al. New plasmid-mediated fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate[J]. Antimicrob Agents Chemother, 2007, 51(9): 3354–3360. DOI: 10.1128/AAC.00339-07 |
| [51] | HENG J, ZHAO Y, LIU M, et al. Substrate-bound structure of the E. coli multidrug resistance transporter MdfA[J]. Cell Res, 2015, 25(9): 1060–1073. DOI: 10.1038/cr.2015.94 |
| [52] | LU S, ZGURSKAYA H I. MacA, a periplasmic membrane fusion protein of the macrolide transporter MacAB-TolC, binds lipopolysaccharide core specifically and with high affinity[J]. J Bacteriol, 2013, 195(21): 4865–4872. DOI: 10.1128/JB.00756-13 |
| [53] | ROUQUETTE-LOUGHLIN C E, BALTHAZAR J T, SHAFER W M. Characterization of the MacA-MacB efflux system in Neisseria gonorrhoeae[J]. J Antimicrob Chemother, 2005, 56(5): 856–860. DOI: 10.1093/jac/dki333 |
| [54] | LEE E H, HILL S A, NAPIER R, et al. Integration Host Factor is required for FarR repression of the farAB-encoded efflux pump of Neisseria gonorrhoeae[J]. Mol Microbiol, 2006, 60(6): 1381–1400. DOI: 10.1111/mmi.2006.60.issue-6 |
| [55] | LEUNG Y M, HOLDBROOK D A, PIGGOT T J, et al. The NorM MATE transporter from N. gonorrhoeae:Insights into drug and ion binding from atomistic molecular dynamics simulations[J]. Biophys J, 2014, 107(2): 460–468. DOI: 10.1016/j.bpj.2014.06.005 |
| [56] | LONG F, ROUQUETTE-LOUGHLIN C, SHAFER W M, et al. Functional cloning and characterization of the multidrug efflux pumps NorM from Neisseria gonorrhoeae and YdhE from Escherichia coli[J]. Antimicrob Agents Chemother, 2008, 52(9): 3052–3060. DOI: 10.1128/AAC.00475-08 |
| [57] | COSTA S S, VIVEIROS M, AMARAL L, et al. Multidrug efflux pumps in Staphylococcus aureus:An update[J]. Open Microbiol J, 2013, 7: 59–71. DOI: 10.2174/1874285801307010059 |
| [58] | TRUONG-BOLDUC Q C, DUNMAN P M, EIDEM T, et al. Transcriptional profiling analysis of the global regulator NorG, a GntR-like protein of Staphylococcus aureus[J]. J Bacteriol, 2011, 193(22): 6207–6214. DOI: 10.1128/JB.05847-11 |
| [59] | TRUONG-BOLDUC Q C, HOOPER D C. Phosphorylation of MgrA and its effect on expression of the NorA and NorB efflux pumps of Staphylococcus aureus[J]. J Bacteriol, 2010, 192(10): 2525–2534. DOI: 10.1128/JB.00018-10 |
| [60] | WALKER J N, CROSBY H A, SPAULDING A R, et al. The Staphylococcus aureus ArlRS two-component system is a novel regulator of agglutination and pathogenesis[J]. PLoS Pathog, 2013, 9(12): e1003819. DOI: 10.1371/journal.ppat.1003819 |
| [61] | BIRUKOU I, SEO S M, SCHINDLER B D, et al. Structural mechanism of transcription regulation of the Staphylococcus aureus multidrug efflux operon mepRA by the MarR family repressor MepR[J]. Nucleic Acids Res, 2014, 42(4): 2774–2788. DOI: 10.1093/nar/gkt1215 |
| [62] | CHOUCHANI C, EL SALABI A, MARRAKCHI R, et al. First report of mefA and msrA/msrB multidrug efflux pumps associated with blaTEM-1 β-lactamase in Enterococcus faecalis[J]. Int J Infect Dis, 2012, 16(2): e104–109. DOI: 10.1016/j.ijid.2011.09.024 |
| [63] | SCHWAIGER K, HÖLZEL C, BAUER J. Detection of the macrolide-efflux protein A gene mef(A) in Enterococcus faecalis[J]. Microb Drug Resist, 2011, 17(3): 429–432. DOI: 10.1089/mdr.2010.0192 |
| [64] | CHANCEY S T, BAI X H, KUMAR N, et al. Transcriptional attenuation controls macrolide inducible efflux and resistance in Streptococcus pneumoniae and in other Gram-positive bacteria containing mef/mel(msr(D)) elements[J]. PLoS One, 2015, 10(2): e0116254. DOI: 10.1371/journal.pone.0116254 |
| [65] | CHANCEY S T, ZHOU X L, ZÄHNER D, et al. Induction of efflux-mediated macrolide resistance in Streptococcus pneumoniae[J]. Antimicrob Agents Chemother, 2011, 55(7): 3413–3422. DOI: 10.1128/AAC.00060-11 |
| [66] | EL GARCH F, LISMOND A, PIDDOCK L J V, et al. Fluoroquinolones induce the expression of patA and patB, which encode ABC efflux pumps in Streptococcus pneumoniae[J]. J Antimicrob Chemother, 2010, 65(10): 2076–2082. DOI: 10.1093/jac/dkq287 |
| [67] | BAYLAY A J, PIDDOCK L J V. Clinically relevant fluoroquinolone resistance due to constitutive overexpression of the PatAB ABC transporter in Streptococcus pneumoniae is conferred by disruption of a transcriptional attenuator[J]. J Antimicrob Chemother, 2015, 70(3): 670–679. DOI: 10.1093/jac/dku449 |
| [68] | HUILLET E, VELGE P, VALLAEYS T, et al. LadR, a new PadR-related transcriptional regulator from Listeria monocytogenes, negatively regulates the expression of the multidrug efflux pump MdrL[J]. FEMS Microbiol Lett, 2006, 254(1): 87–94. DOI: 10.1111/fml.2006.254.issue-1 |
| [69] | JIANG X B, ZHOU L J, GAO D W, et al. Expression of efflux pump gene lde in ciprofloxacin-resistant foodborne isolates of Listeria monocytogenes[J]. Microbiol Immunol, 2012, 56(12): 843–846. DOI: 10.1111/mim.2012.56.issue-12 |
| [70] | GUÉRIN F, GALIMAND M, TUAMBILANGANA F, et al. Overexpression of the novel MATE fluoroquinolone efflux pump FepA in Listeria monocytogenes is driven by inactivation of its local repressor FepR[J]. PLoS One, 2014, 9(9): e106340. DOI: 10.1371/journal.pone.0106340 |
| [71] | PUTMAN M, VAN VEEN H W, DEGENER J E, et al. The lactococcal secondary multidrug transporter LmrP confers resistance to lincosamides, macrolides, streptogramins and tetracyclines[J]. Microbiology, 2001, 147(Pt 10): 2873–2880. |
| [72] | ESPINOSA-URGEL M, SERRANO L, RAMOS J L, et al. Engineering biological approaches for detection of toxic compounds:A new microbial biosensor based on the Pseudomonas putida TtgR repressor[J]. Mol Biotechnol, 2015, 57(6): 558–564. DOI: 10.1007/s12033-015-9849-2 |
| [73] | HONG H, PARK W. TetR repressor-based bioreporters for the detection of doxycycline using Escherichia coli and Acinetobacter oleivorans[J]. Appl Microbiol Biotechnol, 2014, 98(11): 5039–5050. DOI: 10.1007/s00253-014-5566-1 |
| [74] | MELAMED S, NAFTALY S, BELKIN S. Improved detection of antibiotic compounds by bacterial reporter strains achieved by manipulations of membrane permeability and efflux capacity[J]. Appl Microbiol Biotechnol, 2014, 98(5): 2267–2277. DOI: 10.1007/s00253-013-5176-3 |
| [75] | HANSEN L H, FERRARI B, SØRENSEN A H, et al. Detection of oxytetracycline production by Streptomyces rimosus in soil microcosms by combining whole-cell biosensors and flow cytometry[J]. Appl Environ Microbiol, 2001, 67(1): 239–244. DOI: 10.1128/AEM.67.1.239-244.2001 |



